
Introduction
collapse is a C/C++ based package for data transformation and statistical computing in R. It was first released on CRAN end of March 2020. The current version 1.3.1 is a mature piece of statistical software tested with > 7700 unit tests. collapse has 2 main aims:
To facilitate complex data transformation, exploration and computing tasks in R.
(In particular grouped and weighted statistical computations, advanced aggregation of multi-type data, advanced transformations of time series and panel data, and the manipulation of lists)
To help make R code fast, flexible, parsimonious and programmer friendly.
(Provide order of magnitude performance improvements via C/C++ and highly optimized R code, broad object orientation and attribute preservation, and a flexible programming infrastructure in standard and non-standard evaluation)
It is made compatible with dplyr, data.table and the plm approach to panel data. It can be installed in R using:
install.packages('collapse')
# See Documentation
help('collapse-documentation')With this post I want to formally and briefly introduce collapse, provide a basic demonstration of important features, and end with a small benchmark comparing collapse to dplyr and data.table. I hope to convince that collapse provides a superior architecture for data manipulation and statistical computing in R, particularly in terms of flexibility, functionality, performance, and programmability.
Note: Please read this article here for better code appearance.
Demonstration
I start by briefly demonstrating the Fast Statistical Functions, which are a central feature of collapse. Currently there are 14 of them (fmean, fmedian, fmode, fsum, fprod, fsd, fvar, fmin, fmax, fnth, ffirst, flast, fNobs and fNdistinct), they are all S3 generic and support fast grouped and weighted computations on vectors, matrices, data frames, lists and grouped tibbles (class grouped_df). Calling these functions on different objects yields column-wise statistical computations:
library(collapse)
data("iris") # iris dataset in base R
v <- iris$Sepal.Length # Vector
d <- num_vars(iris) # Saving numeric variables
g <- iris$Species # Grouping variable (could also be a list of variables)
# Simple statistics
fmean(v) # Vector
## [1] 5.843333
fsd(qM(d)) # Matrix (qM is a faster as.matrix)
## Sepal.Length Sepal.Width Petal.Length Petal.Width
## 0.8280661 0.4358663 1.7652982 0.7622377
fmode(d) # Data frame
## Sepal.Length Sepal.Width Petal.Length Petal.Width
## 5.0 3.0 1.5 0.2
# Preserving data structure
fmean(qM(d), drop = FALSE) # Still a matrix
## Sepal.Length Sepal.Width Petal.Length Petal.Width
## [1,] 5.843333 3.057333 3.758 1.199333
fmax(d, drop = FALSE) # Still a data.frame
## Sepal.Length Sepal.Width Petal.Length Petal.Width
## 1 7.9 4.4 6.9 2.5The functions fmean, fmedian, fmode, fnth, fsum, fprod, fvar and fsd additionally support weights1.
# Weighted statistics, similarly for vectors and matrices ...
wt <- abs(rnorm(fnrow(iris)))
fmedian(d, w = wt)
## Sepal.Length Sepal.Width Petal.Length Petal.Width
## 5.7 3.0 4.1 1.3The second argument of these functions is called g and supports vectors or lists of grouping variables for grouped computations. For functions supporting weights, w is the third argument2.
# Grouped statistics
fmean(d, g)
## Sepal.Length Sepal.Width Petal.Length Petal.Width
## setosa 5.006 3.428 1.462 0.246
## versicolor 5.936 2.770 4.260 1.326
## virginica 6.588 2.974 5.552 2.026
# Groupwise-weighted statistics
fmean(d, g, wt)
## Sepal.Length Sepal.Width Petal.Length Petal.Width
## setosa 4.964652 3.389885 1.436666 0.2493647
## versicolor 5.924013 2.814171 4.255227 1.3273743
## virginica 6.630702 2.990253 5.601473 2.0724544
fmode(d, g, wt, ties = "max") # Grouped & weighted maximum mode..
## Sepal.Length Sepal.Width Petal.Length Petal.Width
## setosa 5.0 3 1.4 0.2
## versicolor 5.8 3 4.5 1.3
## virginica 6.3 3 5.1 2.3Grouping becomes more efficient when factors or grouping objects are passed to g. Factors can efficiently be created using the function qF, and grouping objects are efficiently created with the function GRP3. As a final layer of complexity, all functions support transformations through the TRA argument.
library(magrittr) # Pipe operators
# Simple Transformations
fnth(v, 0.9, TRA = "replace") %>% head # Replacing values with the 90th percentile
## [1] 6.9 6.9 6.9 6.9 6.9 6.9
fsd(v, TRA = "/") %>% head # Dividing by the overall standard-deviation (scaling)
## [1] 6.158928 5.917402 5.675875 5.555112 6.038165 6.521218
# Grouped transformations
fsd(v, g, TRA = "/") %>% head # Grouped scaling
## [1] 14.46851 13.90112 13.33372 13.05003 14.18481 15.31960
fmin(v, g, TRA = "-") %>% head # Setting the minimum value in each species to 0
## [1] 0.8 0.6 0.4 0.3 0.7 1.1
fsum(v, g, TRA = "%") %>% head # Computing percentages
## [1] 2.037555 1.957651 1.877747 1.837795 1.997603 2.157411
ffirst(v, g, TRA = "%%") %>% head # Taking modulus of first group-value, etc ...
## [1] 0.0 4.9 4.7 4.6 5.0 0.3
# Grouped and weighted transformations
fmedian(v, g, wt, "-") %>% head # Subtracting weighted group-medians
## [1] 0.1 -0.1 -0.3 -0.4 0.0 0.4
fmode(d, g, wt, "replace", ties = "min") %>% head(3) # replace with weighted minimum mode
## Sepal.Length Sepal.Width Petal.Length Petal.Width
## 1 5 3 1.4 0.2
## 2 5 3 1.4 0.2
## 3 5 3 1.4 0.2Currently there are 10 different replacing or sweeping operations supported by TRA, see ?TRA. TRA can also be called directly as a function which performs simple and grouped replacing and sweeping operations with computed statistics:
# Same as fmedian(v, TRA = "-")
TRA(v, median(v), "-") %>% head
## [1] -0.7 -0.9 -1.1 -1.2 -0.8 -0.4
# Replace values with 5% percentile by species
TRA(d, BY(d, g, quantile, 0.05), "replace", g) %>% head(3)
## Sepal.Length Sepal.Width Petal.Length Petal.Width
## 1 4.4 3 1.2 0.1
## 2 4.4 3 1.2 0.1
## 3 4.4 3 1.2 0.1The function BY is generic for Split-Apply-Combine computing with user-supplied functions. Another useful function is dapply (data-apply) for efficient column- and row-operations on matrices and data frames.
Some common panel data transformations like between- and (quasi-)within-transformations (averaging and centering using the mean) are implemented slightly more memory efficient in the functions fbetween and fwithin. The function fscale also exists for fast (grouped, weighted) scaling and centering (standardizing) and mean-preserving scaling. These functions provide further options for data harmonization, such as centering on the overall data mean or scaling to the within-group standard deviation4 (as shown below), as well as scaling / centering to arbitrary supplied means and standard deviations.
oldpar <- par(mfrow = c(1,3))
gv(d, 1:2) %>% { # gv = shortcut for get_vars is > 2x faster than [.data.frame
plot(., col = g, main = "Raw Data")
plot(fwithin(., g, mean = "overall.mean"), col = g,
main = "Centered on Overall Mean")
plot(fscale(., g, mean = "overall.mean", sd = "within.sd"), col = g,
main = "Harmonized Mean and Variance")
}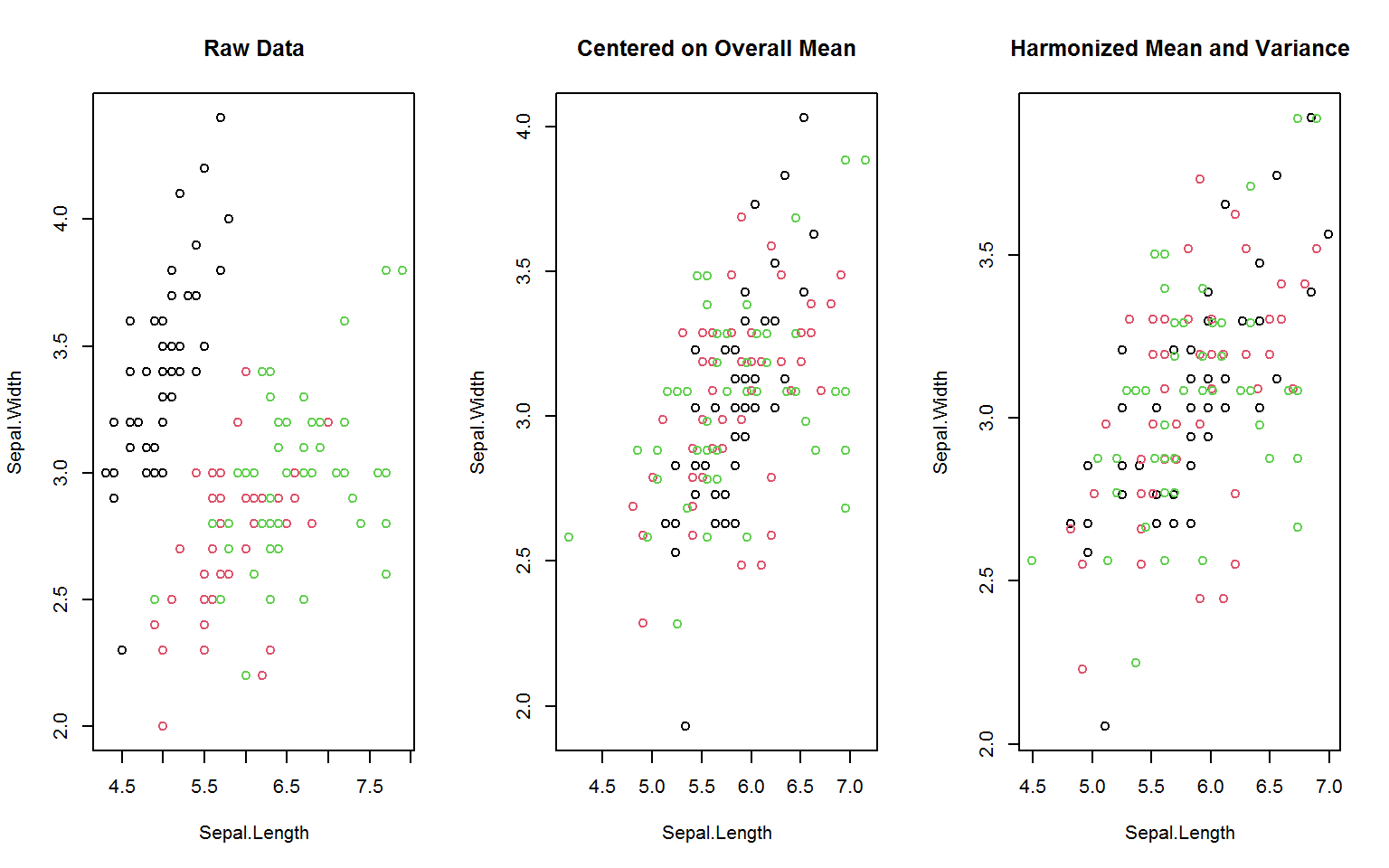
par(oldpar)For the manipulation of time series and panel series, collapse offers the functions flag, fdiff and fgrowth.
# A sequence of lags and leads
flag(EuStockMarkets, -1:1) %>% head(3)
## F1.DAX DAX L1.DAX F1.SMI SMI L1.SMI F1.CAC CAC L1.CAC F1.FTSE FTSE L1.FTSE
## [1,] 1613.63 1628.75 NA 1688.5 1678.1 NA 1750.5 1772.8 NA 2460.2 2443.6 NA
## [2,] 1606.51 1613.63 1628.75 1678.6 1688.5 1678.1 1718.0 1750.5 1772.8 2448.2 2460.2 2443.6
## [3,] 1621.04 1606.51 1613.63 1684.1 1678.6 1688.5 1708.1 1718.0 1750.5 2470.4 2448.2 2460.2
# First and second annual difference of SAX and SMI indices (.c is for non-standard concatenation)
EuStockMarkets[, .c(DAX, SMI)] %>%
fdiff(0:1 * frequency(.), 1:2) %>%
plot(main = c("DAX and SMI")) 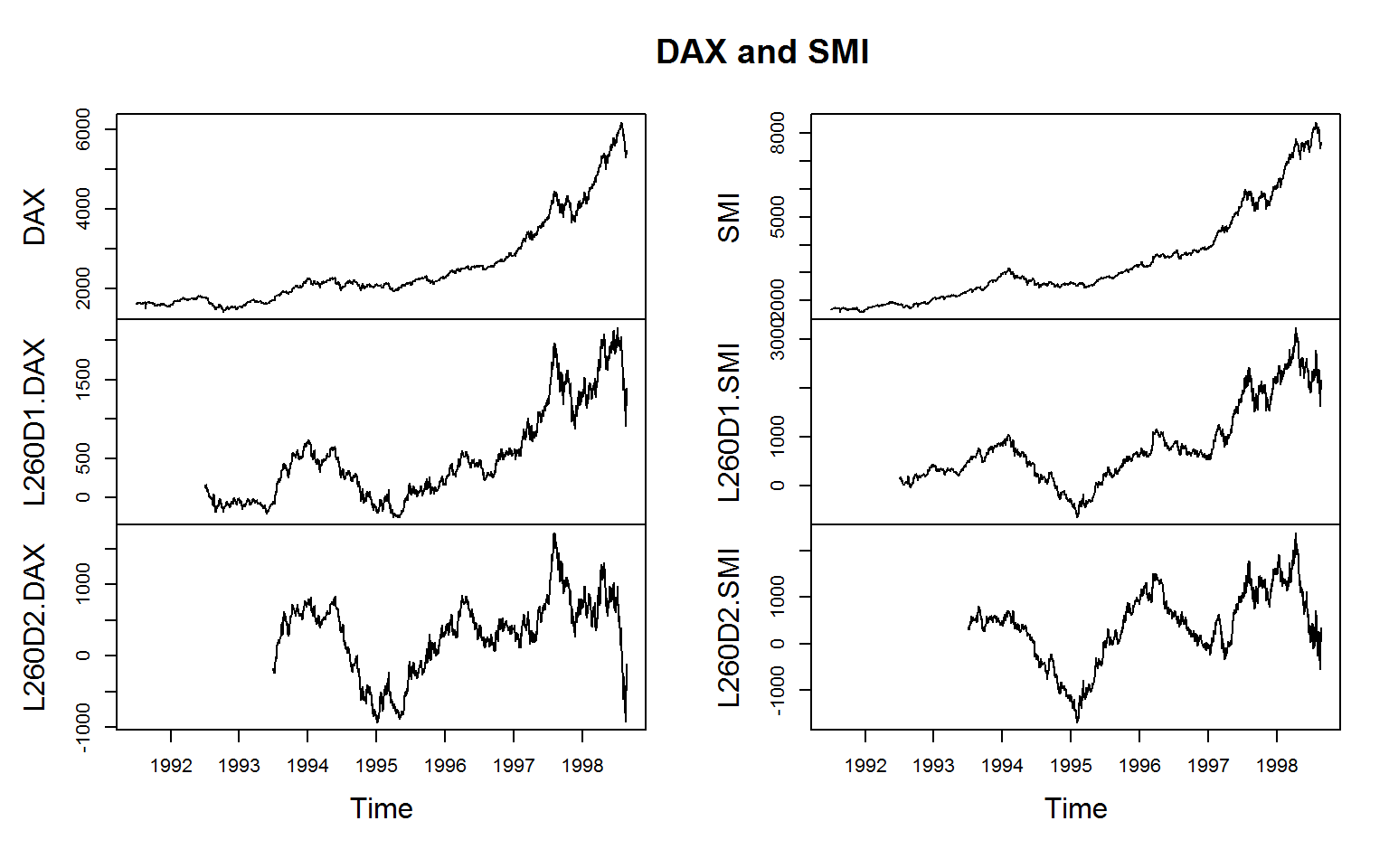
To facilitate programming and integration with dplyr, all functions introduced so far have a grouped_df method.
library(dplyr)
iris %>% add_vars(wt) %>% # Adding weight vector to dataset
filter(Sepal.Length < fmean(Sepal.Length)) %>%
select(Species, Sepal.Width:wt) %>%
group_by(Species) %>% # Frequency-weighted group-variance, default (keep.w = TRUE)
fvar(wt) %>% arrange(sum.wt) # also saves group weights in a column called 'sum.wt'
## # A tibble: 3 x 5
## Species sum.wt Sepal.Width Petal.Length Petal.Width
## <fct> <dbl> <dbl> <dbl> <dbl>
## 1 virginica 3.68 0.0193 0.00993 0.0281
## 2 versicolor 19.2 0.0802 0.181 0.0299
## 3 setosa 43.8 0.142 0.0281 0.0134Since dplyr operations are rather slow, collapse provides its own set of manipulation verbs yielding significant performance gains.
# Same as above.. executes about 15x faster
iris %>% add_vars(wt) %>%
fsubset(Sepal.Length < fmean(Sepal.Length),
Species, Sepal.Width:wt) %>%
fgroup_by(Species) %>%
fvar(wt) %>% roworder(sum.wt)
## # A tibble: 3 x 5
## Species sum.wt Sepal.Width Petal.Length Petal.Width
## <fct> <dbl> <dbl> <dbl> <dbl>
## 1 virginica 3.68 0.0193 0.00993 0.0281
## 2 versicolor 19.2 0.0802 0.181 0.0299
## 3 setosa 43.8 0.142 0.0281 0.0134
# Weighted demeaning
iris %>% fgroup_by(Species) %>% num_vars %>%
fwithin(wt) %>% head(3)
## # A tibble: 3 x 4
## Sepal.Length Sepal.Width Petal.Length Petal.Width
## <dbl> <dbl> <dbl> <dbl>
## 1 0.135 0.110 -0.0367 -0.0494
## 2 -0.0647 -0.390 -0.0367 -0.0494
## 3 -0.265 -0.190 -0.137 -0.0494
# Generate some additional logical data
settransform(iris,
AWMSL = Sepal.Length > fmedian(Sepal.Length, w = wt),
AGWMSL = Sepal.Length > fmedian(Sepal.Length, Species, wt, "replace"))
# Grouped and weighted statistical mode
iris %>% fgroup_by(Species) %>% fmode(wt)
## # A tibble: 3 x 7
## Species Sepal.Length Sepal.Width Petal.Length Petal.Width AWMSL AGWMSL
## <fct> <dbl> <dbl> <dbl> <dbl> <lgl> <lgl>
## 1 setosa 5 3 1.4 0.2 FALSE FALSE
## 2 versicolor 5.8 3 4.5 1.3 TRUE FALSE
## 3 virginica 6.3 3 5.1 2.3 TRUE FALSETo take things a bit further, let’s consider some multilevel / panel data:
# World Bank World Development Data - supplied with collapse
head(wlddev, 3)
## country iso3c date year decade region income OECD PCGDP LIFEEX GINI ODA
## 1 Afghanistan AFG 1961-01-01 1960 1960 South Asia Low income FALSE NA 32.292 NA 114440000
## 2 Afghanistan AFG 1962-01-01 1961 1960 South Asia Low income FALSE NA 32.742 NA 233350000
## 3 Afghanistan AFG 1963-01-01 1962 1960 South Asia Low income FALSE NA 33.185 NA 114880000All variables in this data have labels stored in a ‘label’ attribute (the default if you import from STATA / SPSS / SAS with haven). Variable labels can be accessed and set using vlabels and vlabels<-, and viewed together with names and classes using namlab. In general variable labels and other attributes will be preserved in when working with collapse. collapse provides some of the fastest and most advanced summary statistics:
# Fast distinct value count
fNdistinct(wlddev)
## country iso3c date year decade region income OECD PCGDP LIFEEX GINI ODA
## 216 216 59 59 7 7 4 2 8995 10048 363 7564
# Use descr(wlddev) for a detailed description of each variable
# Checking for within-country variation
varying(wlddev, ~ iso3c)
## country date year decade region income OECD PCGDP LIFEEX GINI ODA
## FALSE TRUE TRUE TRUE FALSE FALSE FALSE TRUE TRUE TRUE TRUE
# Panel data statistics: Summarize GDP and GINI overall, between and within countries
qsu(wlddev, pid = PCGDP + GINI ~ iso3c,
vlabels = TRUE, higher = TRUE)
## , , PCGDP: GDP per capita (constant 2010 US$)
##
## N/T Mean SD Min Max Skew Kurt
## Overall 8995 11563.6529 18348.4052 131.6464 191586.64 3.1121 16.9585
## Between 203 12488.8577 19628.3668 255.3999 141165.083 3.214 17.2533
## Within 44.3103 11563.6529 6334.9523 -30529.0928 75348.067 0.696 17.0534
##
## , , GINI: GINI index (World Bank estimate)
##
## N/T Mean SD Min Max Skew Kurt
## Overall 1356 39.3976 9.6764 16.2 65.8 0.4613 2.2932
## Between 161 39.5799 8.3679 23.3667 61.7143 0.5169 2.6715
## Within 8.4224 39.3976 3.0406 23.9576 54.7976 0.1421 5.7781
# Panel data ACF: Efficient grouped standardizing and computing covariance with panel-lags
psacf(wlddev, ~ iso3c, ~ year, cols = 9:12)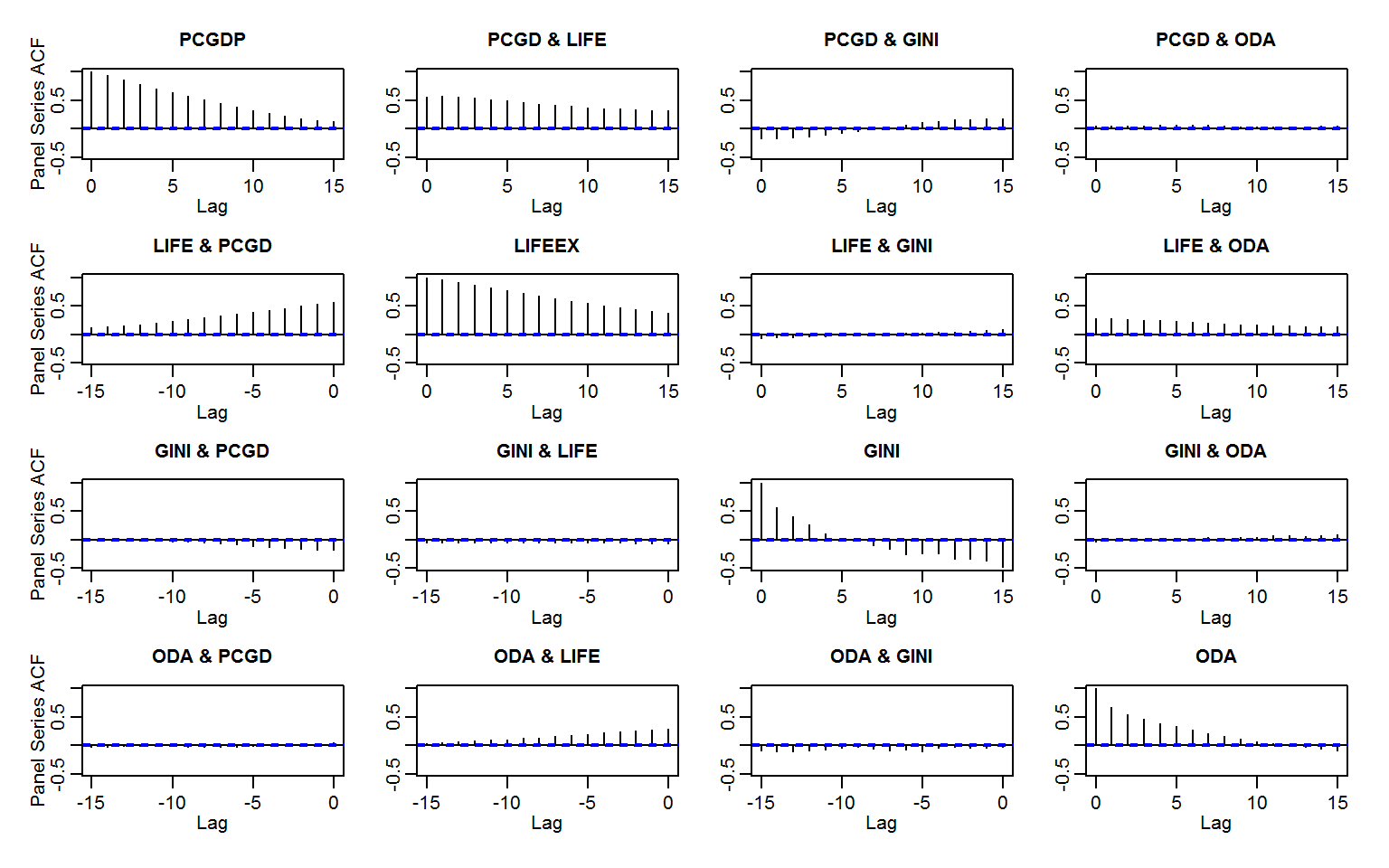
collapse also has its own very flexible data aggregation command called collap, providing fast and easy multi-data-type, multi-function, weighted, parallelized and fully customized data aggregation.
# Applying the mean to numeric and the mode to categorical data (first 2 arguments are 'by' and 'FUN')
collap(wlddev, ~ iso3c + decade, fmean,
catFUN = fmode) %>% head(3)
## country iso3c date year decade region income OECD PCGDP LIFEEX
## 1 Aruba ABW 1961-01-01 1962.5 1960 Latin America & Caribbean High income FALSE NA 66.58583
## 2 Aruba ABW 1967-01-01 1970.0 1970 Latin America & Caribbean High income FALSE NA 69.14178
## 3 Aruba ABW 1976-01-01 1980.0 1980 Latin America & Caribbean High income FALSE NA 72.17600
## GINI ODA
## 1 NA NA
## 2 NA NA
## 3 NA 33630000
# Same as a piped call..
wlddev %>% fgroup_by(iso3c, decade) %>%
collapg(fmean, fmode) %>% head(3)
## # A tibble: 3 x 12
## iso3c decade country date year region income OECD PCGDP LIFEEX GINI ODA
## <fct> <dbl> <chr> <date> <dbl> <fct> <fct> <lgl> <dbl> <dbl> <dbl> <dbl>
## 1 ABW 1960 Aruba 1961-01-01 1962. "Latin America & ~ High inc~ FALSE NA 66.6 NA NA
## 2 ABW 1970 Aruba 1967-01-01 1970 "Latin America & ~ High inc~ FALSE NA 69.1 NA NA
## 3 ABW 1980 Aruba 1976-01-01 1980 "Latin America & ~ High inc~ FALSE NA 72.2 NA 3.36e7
# Same thing done manually... without column reordering
wlddev %>% fgroup_by(iso3c, decade) %>% {
add_vars(fmode(cat_vars(.)), # cat_vars selects non-numeric (categorical) columns
fmean(num_vars(.), keep.group_vars = FALSE))
} %>% head(3)
## # A tibble: 3 x 12
## iso3c decade country date region income OECD year PCGDP LIFEEX GINI ODA
## <fct> <dbl> <chr> <date> <fct> <fct> <lgl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 ABW 1960 Aruba 1961-01-01 "Latin America & ~ High inc~ FALSE 1962. NA 66.6 NA NA
## 2 ABW 1970 Aruba 1967-01-01 "Latin America & ~ High inc~ FALSE 1970 NA 69.1 NA NA
## 3 ABW 1980 Aruba 1976-01-01 "Latin America & ~ High inc~ FALSE 1980 NA 72.2 NA 3.36e7
# Adding weights: weighted mean and weighted mode (catFUN is 3rd argument)
wlddev$weights <- abs(rnorm(fnrow(wlddev)))
collap(wlddev, ~ iso3c + decade, fmean, fmode, # weights are also aggregated using sum
w = ~ weights, wFUN = fsum) %>% head(3)
## country iso3c date year decade region income OECD PCGDP
## 1 Aruba ABW 1965-01-01 1963.375 1960 Latin America & Caribbean High income FALSE NA
## 2 Aruba ABW 1967-01-01 1969.179 1970 Latin America & Caribbean High income FALSE NA
## 3 Aruba ABW 1980-01-01 1980.443 1980 Latin America & Caribbean High income FALSE NA
## LIFEEX GINI ODA weights
## 1 66.87902 NA NA 4.527996
## 2 68.85522 NA NA 7.314234
## 3 72.29649 NA 33630000 6.525710
# Can also apply multiple functions to columns, return in wide or long format or as list of data frames
collap(wlddev, PCGDP + LIFEEX ~ region + income,
list(fmean, fsd, fmin, fmax), return = "long") %>% head(3)
## Function region income PCGDP LIFEEX
## 1 fmean East Asia & Pacific High income 26042.280 73.22799
## 2 fmean East Asia & Pacific Lower middle income 1621.178 58.83796
## 3 fmean East Asia & Pacific Upper middle income 3432.559 66.41750The default (keep.col.order = TRUE) ensures that the data remains in the same order, and, when working with Fast Statistical Functions, all column attributes are preserved. When aggregating with multiple functions, you can parallelize over them (internally done with parallel::mclapply).
Time computations on panel data are also simple and computationally very fast.
# Panel Lag and lead of PCGDP and LIFEEX
L(wlddev, -1:1, PCGDP + LIFEEX ~ iso3c, ~year) %>% head3
## iso3c year F1.PCGDP PCGDP L1.PCGDP F1.LIFEEX LIFEEX L1.LIFEEX
## 1 AFG 1960 NA NA NA 32.742 32.292 NA
## 2 AFG 1961 NA NA NA 33.185 32.742 32.292
## 3 AFG 1962 NA NA NA 33.624 33.185 32.742
# Equivalent piped call
wlddev %>% fgroup_by(iso3c) %>%
fselect(iso3c, year, PCGDP, LIFEEX) %>%
flag(-1:1, year) %>% head(3)
## # A tibble: 3 x 8
## iso3c year F1.PCGDP PCGDP L1.PCGDP F1.LIFEEX LIFEEX L1.LIFEEX
## <fct> <int> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 AFG 1960 NA NA NA 32.7 32.3 NA
## 2 AFG 1961 NA NA NA 33.2 32.7 32.3
## 3 AFG 1962 NA NA NA 33.6 33.2 32.7
# Or using plm classes for panel data
pwlddev <- plm::pdata.frame(wlddev, index = .c(iso3c, year))
L(pwlddev, -1:1, cols = .c(PCGDP, LIFEEX)) %>% head(3)
## iso3c year F1.PCGDP PCGDP L1.PCGDP F1.LIFEEX LIFEEX L1.LIFEEX
## ABW-1960 ABW 1960 NA NA NA 66.074 65.662 NA
## ABW-1961 ABW 1961 NA NA NA 66.444 66.074 65.662
## ABW-1962 ABW 1962 NA NA NA 66.787 66.444 66.074
# Growth rates in percentage terms: 1 and 10-year
G(pwlddev, c(1, 10), cols = 9:12) %>% head(3) # or use Dlog, or G(..., logdiff = TRUE) for percentages
## iso3c year G1.PCGDP L10G1.PCGDP G1.LIFEEX L10G1.LIFEEX G1.GINI L10G1.GINI G1.ODA L10G1.ODA
## ABW-1960 ABW 1960 NA NA NA NA NA NA NA NA
## ABW-1961 ABW 1961 NA NA 0.6274558 NA NA NA NA NA
## ABW-1962 ABW 1962 NA NA 0.5599782 NA NA NA NA NAEquivalently we can can compute lagged / leaded and suitably iterated (log-) differences, as well as quasi-(log-)differences of the form \(x_t - \rho x_{t-1}\). The operators L, D, Dlog and G are shorthand’s for the functions flag, fdiff and fgrowth allowing formula input. Similar operators exist for fwithin, fscale, etc. which also support plm classes.
This short demonstration illustrated some basic features of collapse. A more complete overview of the package is provided in the documentation and the vignettes.
Benchmark
For benchmarking I use some product-level trade data from the UN Comtrade database, processed by tadestatistics.io.
library(tradestatistics)
# US HS4-level trade from 2000 to 2018
us_trade <- ots_create_tidy_data(years = 2000:2018,
reporters = "usa",
table = "yrpc")Downloading US product-level trade (HS4) from 2000 to 2018 gives about 2.6 million observations:
fdim(us_trade)
## [1] 2569787 16
head(us_trade, 1)
## year reporter_iso reporter_fullname_english
## 1: 2017 usa USA, Puerto Rico and US Virgin Islands (excludes Virgin Islands until 1981)
## partner_iso partner_fullname_english section_code section_color section_shortname_english
## 1: afg Afghanistan 01 #74c0e2 Animal Products
## section_fullname_english group_code group_fullname_english product_code
## 1: Live Animals; Animal Products 01 Animals; live 0101
## product_shortname_english product_fullname_english export_value_usd
## 1: Horses Horses, asses, mules and hinnies; live 3005
## import_value_usd
## 1: NA
# 19 years, 221 trading partners, 1222 products, unbalanced panel with product-time gaps...
fNdistinct(us_trade)
## year reporter_iso reporter_fullname_english
## 19 1 1
## partner_iso partner_fullname_english section_code
## 221 221 22
## section_color section_shortname_english section_fullname_english
## 22 22 22
## group_code group_fullname_english product_code
## 97 97 1222
## product_shortname_english product_fullname_english export_value_usd
## 1217 1222 1081492
## import_value_usd
## 684781
# Summarizing data between and within partner-product pairs
qsu(us_trade, pid = export_value_usd + import_value_usd ~ partner_iso + product_code)
## , , export_value_usd
##
## N/T Mean SD Min Max
## Overall 2,450301 11,054800.6 157,295999 1 2.83030606e+10
## Between 205513 7,268011.31 118,709845 1 1.66436161e+10
## Within 11.9229 11,054800.6 68,344396.5 -1.01599067e+10 1.67185229e+10
##
## , , import_value_usd
##
## N/T Mean SD Min Max
## Overall 1,248201 31,421502.4 505,644905 1 8.51970855e+10
## Between 130114 16,250758.2 328,538895 1 4.36545695e+10
## Within 9.5931 31,421502.4 212,076350 -3.32316111e+10 4.15739375e+10It would also be interesting to summarize the trade flows for each partner, but that would be too large to print to the console. We can however get the qsu output as a list of matrices:
# Doing all of that by partner - variance of flows between and within traded products for each partner
l <- qsu(us_trade,
by = export_value_usd + import_value_usd ~ partner_iso,
pid = ~ partner_iso + product_code, array = FALSE)
str(l, give.attr = FALSE)
## List of 2
## $ export_value_usd:List of 3
## ..$ Overall: 'qsu' num [1:221, 1:5] 7250 12427 6692 5941 4017 ...
## ..$ Between: 'qsu' num [1:221, 1:5] 901 1151 872 903 695 ...
## ..$ Within : 'qsu' num [1:221, 1:5] 8.05 10.8 7.67 6.58 5.78 ...
## $ import_value_usd:List of 3
## ..$ Overall: 'qsu' num [1:221, 1:5] 1157 1547 361 1512 685 ...
## ..$ Between: 'qsu' num [1:221, 1:5] 312 532 167 347 235 ...
## ..$ Within : 'qsu' num [1:221, 1:5] 3.71 2.91 2.16 4.36 2.91 ...Now with the function unlist2d, we can efficiently turn this into a tidy data frame:
unlist2d(l, idcols = c("Variable", "Trans"),
row.names = "Partner", DT = TRUE) %>% head(3)
## Variable Trans Partner N Mean SD Min Max
## 1: export_value_usd Overall afg 7250 2170074.0 21176449.3 56 1115125722
## 2: export_value_usd Overall ago 12427 2188174.6 17158413.8 1 687323408
## 3: export_value_usd Overall aia 6692 125729.3 586862.2 2503 17698445If l were some statistical object we could first pull out relevant elements using get_elem, possibly process those elements using rapply2d and then apply unlist2d to get the data frame (or data.table with DT = TRUE). These are the main collapse list-processing functions.
Now on to the benchmark. It is run on a Windows 8.1 laptop with a 2x 2.2 GHZ Intel i5 processor, 8GB DDR3 RAM and a Samsung 850 EVO SSD hard drive.
library(microbenchmark)
library(dplyr)
library(data.table) # Default for this machine is 2 threads
# Grouping (data.table:::forderv does not compute the unique groups yet)
microbenchmark(collapse = fgroup_by(us_trade, partner_iso, group_code, year),
data.table = data.table:::forderv(us_trade, c("partner_iso", "group_code", "year"), retGrp = TRUE),
dplyr = group_by(us_trade, partner_iso, group_code, year), times = 10)
## Unit: milliseconds
## expr min lq mean median uq max neval cld
## collapse 97.23371 98.32389 123.7255 107.0449 135.9751 239.6009 10 a
## data.table 102.88811 107.66742 1267.0891 111.4340 118.1944 11658.7908 10 a
## dplyr 924.67524 979.54759 1042.8808 1008.5981 1078.1861 1354.4466 10 a
# Sum
microbenchmark(collapse = collap(us_trade, export_value_usd + import_value_usd ~ partner_iso + group_code + year, fsum),
data.table = us_trade[, list(export_value_usd = sum(export_value_usd, na.rm = TRUE),
import_value_usd = sum(import_value_usd, na.rm = TRUE)),
by = c("partner_iso", "group_code", "year")],
dplyr = group_by(us_trade, partner_iso, group_code, year) %>%
dplyr::select(export_value_usd, import_value_usd) %>% summarise_all(sum, na.rm = TRUE), times = 10)
## Unit: milliseconds
## expr min lq mean median uq max neval cld
## collapse 107.1917 116.5861 128.3973 118.9365 150.3367 161.3135 10 a
## data.table 169.3562 182.7400 198.2929 189.3267 221.8608 226.7869 10 a
## dplyr 2332.0459 2426.7485 2652.3324 2568.5637 2791.4008 3263.4077 10 b
# Mean
microbenchmark(collapse = collap(us_trade, export_value_usd + import_value_usd ~ partner_iso + group_code + year, fmean),
data.table = us_trade[, list(export_value_usd = mean(export_value_usd, na.rm = TRUE),
import_value_usd = mean(import_value_usd, na.rm = TRUE)),
by = c("partner_iso", "group_code", "year")],
dplyr = group_by(us_trade, partner_iso, group_code, year) %>%
dplyr::select(export_value_usd, import_value_usd) %>% summarise_all(mean, na.rm = TRUE), times = 10)
## Unit: milliseconds
## expr min lq mean median uq max neval cld
## collapse 121.3204 125.5664 142.8944 132.4904 137.1090 214.3933 10 a
## data.table 177.7091 187.3493 210.5166 201.0806 224.2513 269.5401 10 a
## dplyr 6303.0662 7037.0073 7270.0693 7242.0813 7872.5290 8066.6510 10 b
# Variance
microbenchmark(collapse = collap(us_trade, export_value_usd + import_value_usd ~ partner_iso + group_code + year, fvar),
data.table = us_trade[, list(export_value_usd = var(export_value_usd, na.rm = TRUE),
import_value_usd = var(import_value_usd, na.rm = TRUE)),
by = c("partner_iso", "group_code", "year")],
dplyr = group_by(us_trade, partner_iso, group_code, year) %>%
dplyr::select(export_value_usd, import_value_usd) %>% summarise_all(var, na.rm = TRUE), times = 10)
## Unit: milliseconds
## expr min lq mean median uq max neval cld
## collapse 123.8475 126.6842 135.6485 134.5366 142.3431 153.3176 10 a
## data.table 269.8578 284.1863 300.0291 287.6637 300.2628 365.7684 10 a
## dplyr 10408.3787 10815.5928 11298.6246 11225.9573 11726.4892 12275.4899 10 b
# Mode (forget trying to do this with dplyr or data.table using some mode function created in base R, it runs forever...)
microbenchmark(collapse = fgroup_by(us_trade, partner_iso, group_code, year) %>%
fselect(export_value_usd, import_value_usd) %>% fmode, times = 10)
## Unit: milliseconds
## expr min lq mean median uq max neval
## collapse 413.1148 430.7063 451.1648 443.147 459.1362 525.748 10
# Weighted Mean (not easily done with dplyr)
settransform(us_trade, weights = abs(rnorm(length(year))))
microbenchmark(collapse = collap(us_trade, export_value_usd + import_value_usd ~ partner_iso + group_code + year, fmean, w = ~ weights, keep.w = FALSE),
data.table = us_trade[, list(export_value_usd = weighted.mean(export_value_usd, weights, na.rm = TRUE),
import_value_usd = weighted.mean(import_value_usd, weights, na.rm = TRUE)),
by = c("partner_iso", "group_code", "year")], times = 10)
## Unit: milliseconds
## expr min lq mean median uq max neval cld
## collapse 114.2594 115.2599 118.7735 118.5715 122.2374 124.0246 10 a
## data.table 5808.7005 5867.3619 5957.9288 5904.3373 5963.8040 6507.7393 10 b
# Replace values with group-sum
microbenchmark(collapse = fgroup_by(us_trade, partner_iso, group_code, year) %>%
fselect(export_value_usd, import_value_usd) %>% fsum(TRA = "replace_fill"),
data.table = us_trade[, `:=`(export_value_usd2 = sum(export_value_usd, na.rm = TRUE),
import_value_usd2 = sum(import_value_usd, na.rm = TRUE)),
by = c("partner_iso", "group_code", "year")],
dplyr = group_by(us_trade, partner_iso, group_code, year) %>%
dplyr::select(export_value_usd, import_value_usd) %>% mutate_all(sum, na.rm = TRUE), times = 10)
## Unit: milliseconds
## expr min lq mean median uq max neval cld
## collapse 128.8186 139.7133 154.8318 153.3190 167.7827 189.6136 10 a
## data.table 823.7694 849.4982 861.1539 853.3544 869.0394 917.6767 10 b
## dplyr 2464.5943 2653.9227 2812.6778 2772.3155 2884.5482 3335.0273 10 c
# Centering, partner-product
microbenchmark(collapse = fgroup_by(us_trade, partner_iso, product_code) %>%
fselect(export_value_usd, import_value_usd) %>% fwithin,
data.table = us_trade[, `:=`(export_value_usd2 = export_value_usd - mean(export_value_usd, na.rm = TRUE),
import_value_usd2 = import_value_usd - mean(import_value_usd, na.rm = TRUE)),
by = c("partner_iso", "group_code", "year")],
dplyr = group_by(us_trade, partner_iso, group_code, year) %>%
dplyr::select(export_value_usd, import_value_usd) %>% mutate_all(function(x) x - mean(x, na.rm = TRUE)), times = 10)
## Unit: milliseconds
## expr min lq mean median uq max neval cld
## collapse 80.1893 88.50289 96.74396 96.62168 105.6455 109.0914 10 a
## data.table 4537.3741 4578.44447 4788.89139 4816.80366 4876.4886 5071.2853 10 b
## dplyr 6822.4752 7190.71493 7388.08024 7372.80011 7706.3514 7902.9720 10 c
# Lag
# Much better to sort data for dplyr
setorder(us_trade, partner_iso, product_code, year)
# We have an additional problem here: There are time-gaps within some partner-product pairs
tryCatch(L(us_trade, 1, export_value_usd + import_value_usd ~ partner_iso + product_code, ~ year),
error = function(e) e)
## <Rcpp::exception in L.data.frame(us_trade, 1, export_value_usd + import_value_usd ~ partner_iso + product_code, ~year): Gaps in timevar within one or more groups>
# The solution is that we create a unique id for each continuous partner-product sequence
settransform(us_trade, id = seqid(year + unattrib(finteraction(partner_iso, product_code)) * 20L))
# Notes: Normally id = seqid(year) would be enough on sorted data, but here we also have very different start and end dates, with the potential of overlaps...
fNdistinct(us_trade$id)
## [1] 423884
# Another comparison..
microbenchmark(fNdistinct(us_trade$id), n_distinct(us_trade$id))
## Unit: milliseconds
## expr min lq mean median uq max neval cld
## fNdistinct(us_trade$id) 26.46380 26.84445 27.91192 27.25768 28.42417 37.60704 100 a
## n_distinct(us_trade$id) 56.70598 64.67573 69.11154 65.50040 68.60963 145.39718 100 b
# Here we go now:
microbenchmark(collapse = L(us_trade, 1, export_value_usd + import_value_usd ~ id),
collapse_ordered = L(us_trade, 1, export_value_usd + import_value_usd ~ id, ~ year),
data.table = us_trade[, shift(.SD), keyby = id,
.SDcols = c("export_value_usd","import_value_usd")],
data.table_ordered = us_trade[order(year), shift(.SD), keyby = id,
.SDcols = c("export_value_usd","import_value_usd")],
dplyr = group_by(us_trade, id) %>% dplyr::select(export_value_usd, import_value_usd) %>%
mutate_all(lag), times = 10)
## Unit: milliseconds
## expr min lq mean median uq max neval
## collapse 18.57103 29.47686 34.75352 34.34208 39.30635 55.87373 10
## collapse_ordered 51.26266 57.90416 67.40628 66.86212 73.63482 94.11623 10
## data.table 7594.63120 7820.26944 7968.29873 7879.71628 8266.03880 8353.21097 10
## data.table_ordered 7622.66044 7635.12055 8090.12031 7726.25161 8492.04252 9428.44336 10
## dplyr 32428.73832 32583.82844 33584.48046 32903.50014 34725.72392 36189.46410 10
## cld
## a
## a
## b
## b
## c
# Note: you can do ordered lags using mutate_all(lag, order_by = "year") for dplyr, but at computation times in excess of 90 seconds..
The benchmarks show that collapse is consistently very fast. More extensive benchmarks against dplyr and plm are provided in the corresponding vignettes.
fvarandfsdcompute frequency weights, the most common form of weighted sample variance. ↩︎I note that all further examples generalize to different objects (vectors, matrices, data frames).↩︎
Grouping objects are better for programming and for multiple grouping variables. This is demonstrated in the blog post on programming with collapse.↩︎
The within-group standard deviation is the standard deviation computed on the group-centered data.↩︎