
Auto- and Cross- Covariance and Correlation Function Estimation for Panel Series
psacf.Rdpsacf, pspacf and psccf compute (and by default plot) estimates of the auto-, partial auto- and cross- correlation or covariance functions for panel series. They are analogues to acf, pacf and ccf.
Usage
psacf(x, ...)
pspacf(x, ...)
psccf(x, y, ...)
# Default S3 method
psacf(x, g, t = NULL, lag.max = NULL, type = c("correlation", "covariance","partial"),
plot = TRUE, gscale = TRUE, ...)
# Default S3 method
pspacf(x, g, t = NULL, lag.max = NULL, plot = TRUE, gscale = TRUE, ...)
# Default S3 method
psccf(x, y, g, t = NULL, lag.max = NULL, type = c("correlation", "covariance"),
plot = TRUE, gscale = TRUE, ...)
# S3 method for class 'data.frame'
psacf(x, by, t = NULL, cols = is.numeric, lag.max = NULL,
type = c("correlation", "covariance","partial"), plot = TRUE, gscale = TRUE, ...)
# S3 method for class 'data.frame'
pspacf(x, by, t = NULL, cols = is.numeric, lag.max = NULL,
plot = TRUE, gscale = TRUE, ...)
# Methods for indexed data / compatibility with plm:
# S3 method for class 'pseries'
psacf(x, lag.max = NULL, type = c("correlation", "covariance","partial"),
plot = TRUE, gscale = TRUE, ...)
# S3 method for class 'pseries'
pspacf(x, lag.max = NULL, plot = TRUE, gscale = TRUE, ...)
# S3 method for class 'pseries'
psccf(x, y, lag.max = NULL, type = c("correlation", "covariance"),
plot = TRUE, gscale = TRUE, ...)
# S3 method for class 'pdata.frame'
psacf(x, cols = is.numeric, lag.max = NULL,
type = c("correlation", "covariance","partial"), plot = TRUE, gscale = TRUE, ...)
# S3 method for class 'pdata.frame'
pspacf(x, cols = is.numeric, lag.max = NULL, plot = TRUE, gscale = TRUE, ...)Arguments
- x, y
a numeric vector, 'indexed_series' ('pseries'), data frame or 'indexed_frame' ('pdata.frame').
- g
a factor,
GRPobject, or atomic vector / list of vectors (internally grouped withgroup) used to groupx.- by
data.frame method: Same input as
g, but also allows one- or two-sided formulas using the variables inx, i.e.~ idvarorvar1 + var2 ~ idvar1 + idvar2.- t
a time vector or list of vectors. See
flag.- cols
data.frame method: Select columns using a function, column names, indices or a logical vector. Note:
colsis ignored if a two-sided formula is passed toby.- lag.max
integer. Maximum lag at which to calculate the acf. Default is
2*sqrt(length(x)/ng)wherengis the number of groups in the panel series / supplied tog.- type
character. String giving the type of acf to be computed. Allowed values are "correlation" (the default), "covariance" or "partial".
- plot
logical. If
TRUE(default) the acf is plotted.- gscale
logical. Do a groupwise scaling / standardization of
x, y(usingfscaleand the groups supplied tog) before computing panel-autocovariances / correlations. See Details.- ...
further arguments to be passed to
plot.acf.
Details
If gscale = TRUE data are standardized within each group (using fscale) such that the group-mean is 0 and the group-standard deviation is 1. This is strongly recommended for most panels to get rid of individual-specific heterogeneity which would corrupt the ACF computations.
After scaling, psacf, pspacf and psccf compute the ACF/CCF by creating a matrix of panel-lags of the series using flag and then computing the covariance of this matrix with the series (x, y) using cov and pairwise-complete observations, and dividing by the variance (of x, y). Creating the lag matrix may require a lot of memory on large data, but passing a sequence of lags to flag and thus calling flag and cov one time is generally much faster than calling them lag.max times. The partial ACF is computed from the ACF using a Yule-Walker decomposition, in the same way as in pacf.
Value
An object of class 'acf', see acf. The result is returned invisibly if plot = TRUE.
Examples
## World Development Panel Data
head(wlddev) # See also help(wlddev)
#> country iso3c date year decade region income OECD PCGDP
#> 1 Afghanistan AFG 1961-01-01 1960 1960 South Asia Low income FALSE NA
#> 2 Afghanistan AFG 1962-01-01 1961 1960 South Asia Low income FALSE NA
#> 3 Afghanistan AFG 1963-01-01 1962 1960 South Asia Low income FALSE NA
#> 4 Afghanistan AFG 1964-01-01 1963 1960 South Asia Low income FALSE NA
#> 5 Afghanistan AFG 1965-01-01 1964 1960 South Asia Low income FALSE NA
#> LIFEEX GINI ODA POP
#> 1 32.446 NA 116769997 8996973
#> 2 32.962 NA 232080002 9169410
#> 3 33.471 NA 112839996 9351441
#> 4 33.971 NA 237720001 9543205
#> 5 34.463 NA 295920013 9744781
#> [ reached 'max' / getOption("max.print") -- omitted 1 rows ]
psacf(wlddev$PCGDP, wlddev$country, wlddev$year) # ACF of GDP per Capita
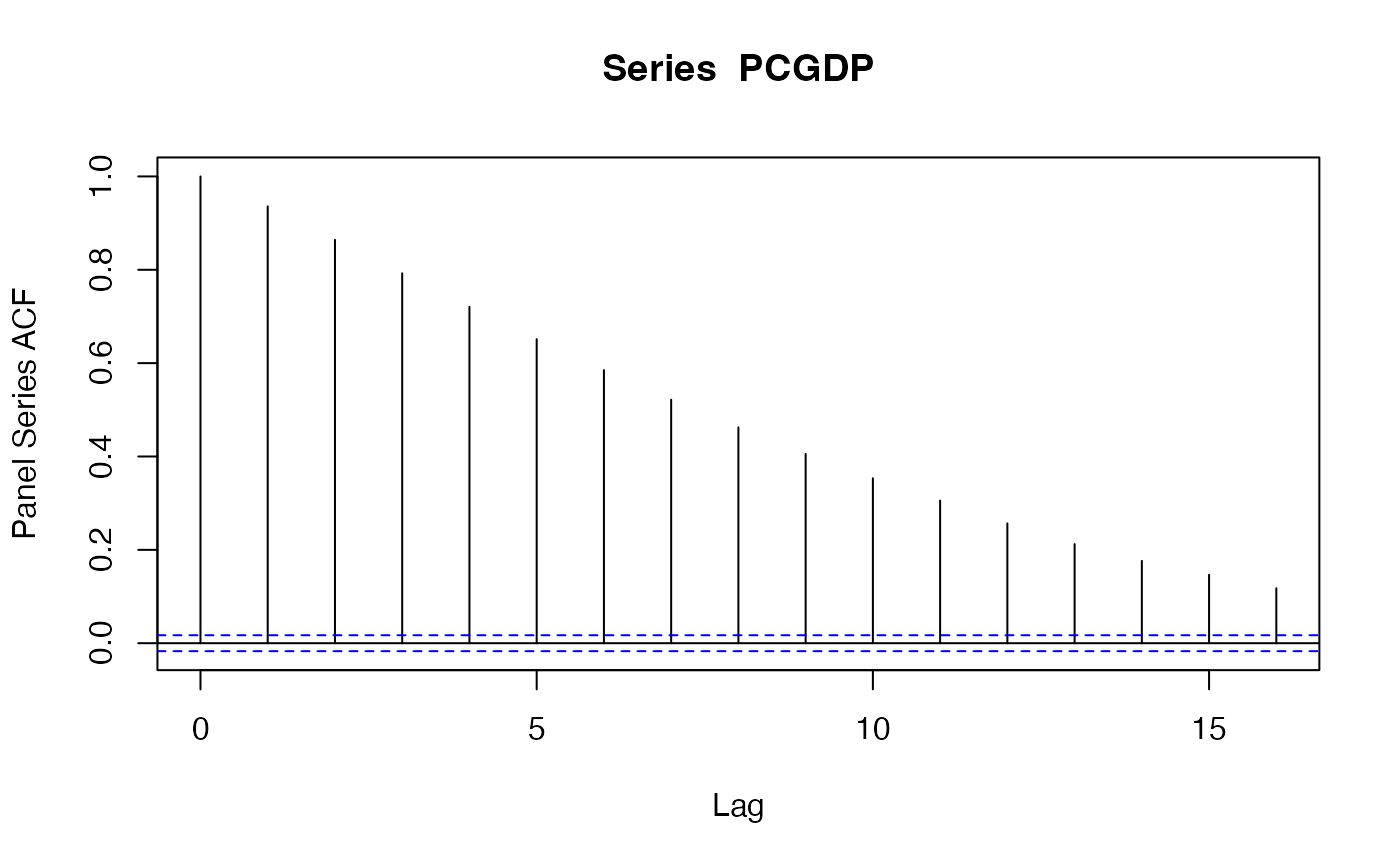 psacf(wlddev, PCGDP ~ country, ~year) # Same using data.frame method
psacf(wlddev, PCGDP ~ country, ~year) # Same using data.frame method
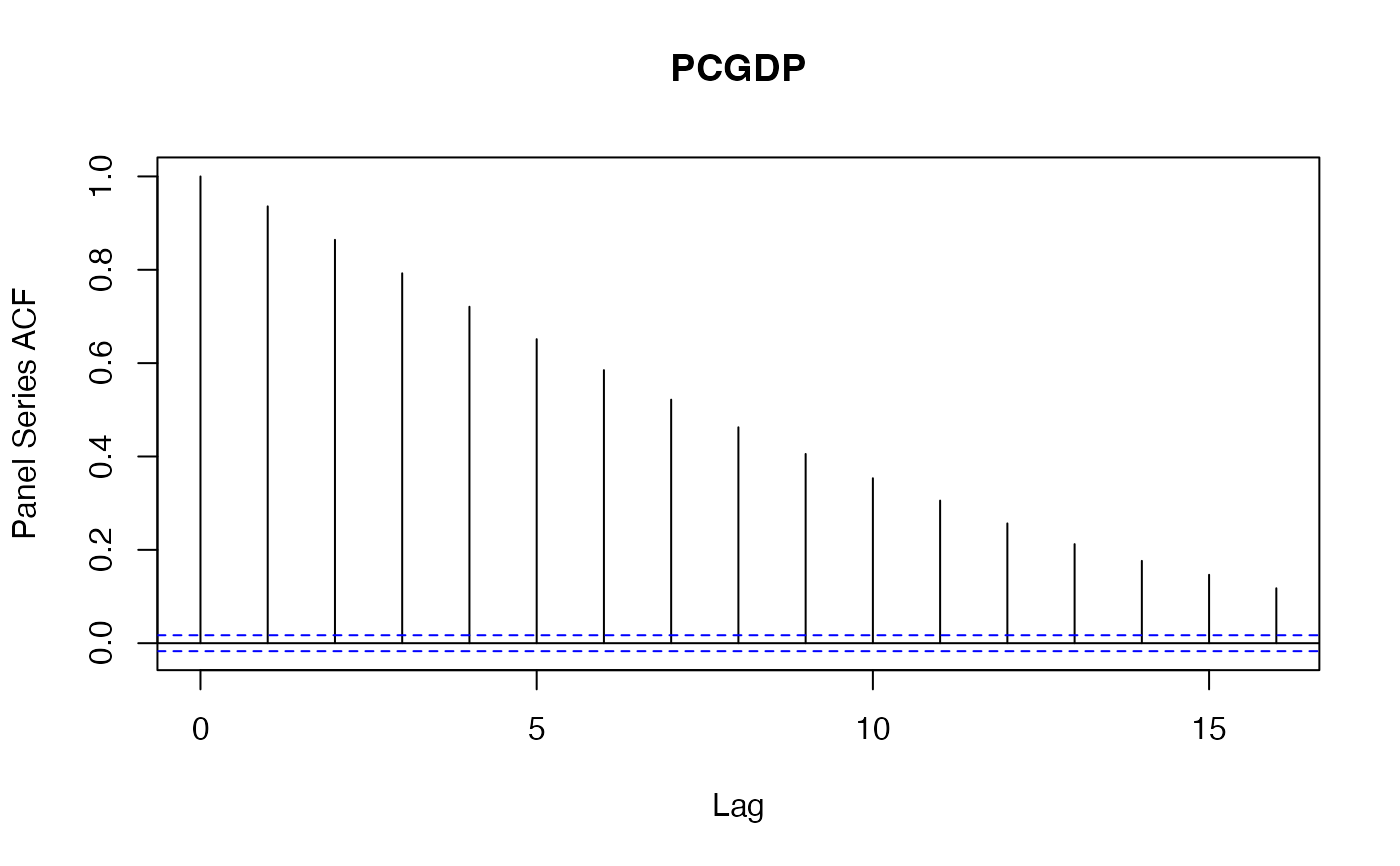 psacf(wlddev$PCGDP, wlddev$country) # The Data is sorted, can omit t
psacf(wlddev$PCGDP, wlddev$country) # The Data is sorted, can omit t
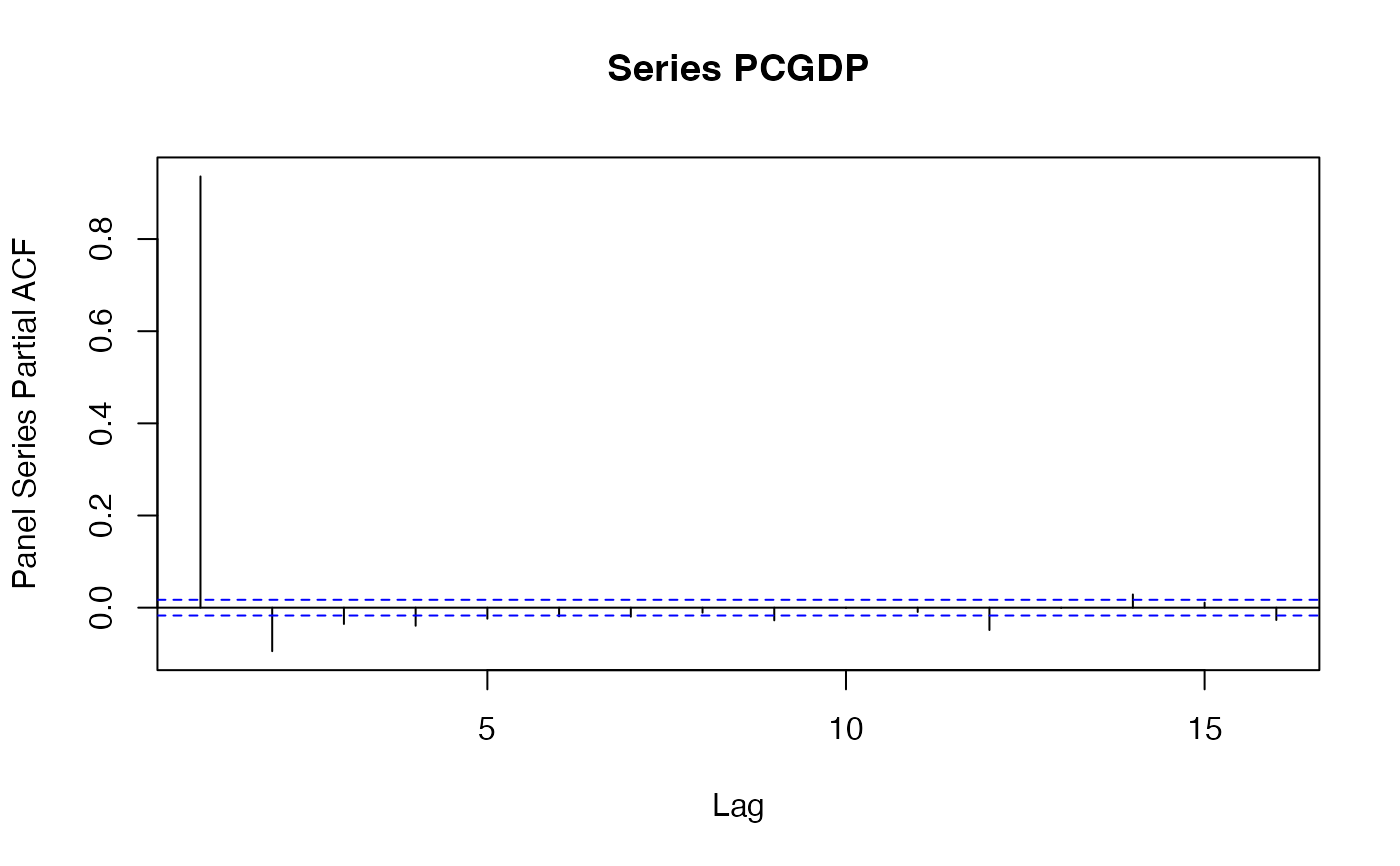
 pspacf(wlddev$PCGDP, wlddev$country) # Partial ACF
pspacf(wlddev$PCGDP, wlddev$country) # Partial ACF
 psccf(wlddev$PCGDP, wlddev$LIFEEX, wlddev$country) # CCF with Life-Expectancy at Birth
psccf(wlddev$PCGDP, wlddev$LIFEEX, wlddev$country) # CCF with Life-Expectancy at Birth
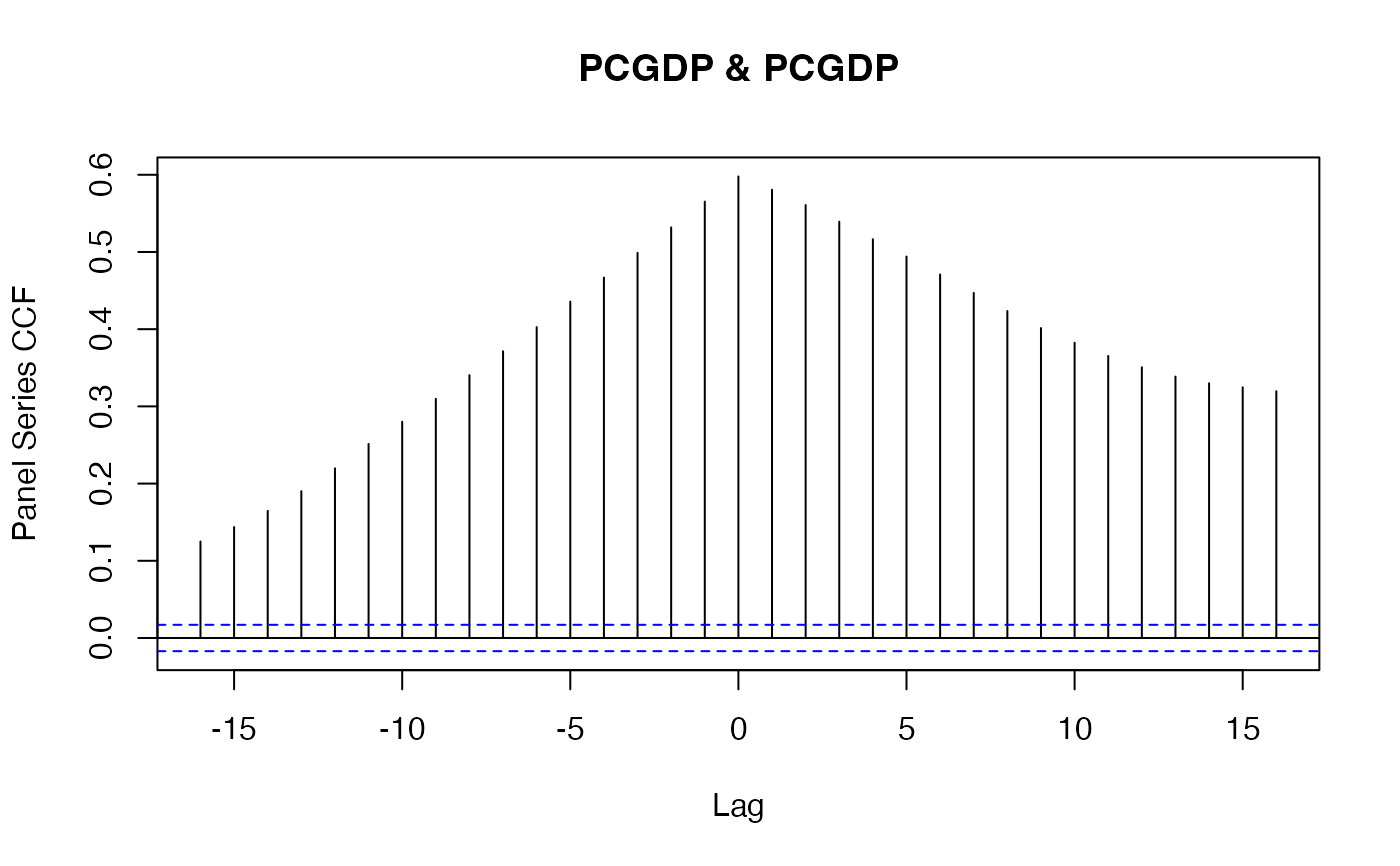 psacf(wlddev, PCGDP + LIFEEX + ODA ~ country, ~year) # ACF and CCF of GDP, LIFEEX and ODA
psacf(wlddev, PCGDP + LIFEEX + ODA ~ country, ~year) # ACF and CCF of GDP, LIFEEX and ODA
 psacf(wlddev, ~ country, ~year, c(9:10,12)) # Same, using cols argument
pspacf(wlddev, ~ country, ~year, c(9:10,12)) # Partial ACF
psacf(wlddev, ~ country, ~year, c(9:10,12)) # Same, using cols argument
pspacf(wlddev, ~ country, ~year, c(9:10,12)) # Partial ACF
 ## Using indexed data:
wldi <- findex_by(wlddev, iso3c, year) # Creating a indexed frame
PCGDP <- wldi$PCGDP # Indexed Series of GDP per Capita
LIFEEX <- wldi$LIFEEX # Indexed Series of Life Expectancy
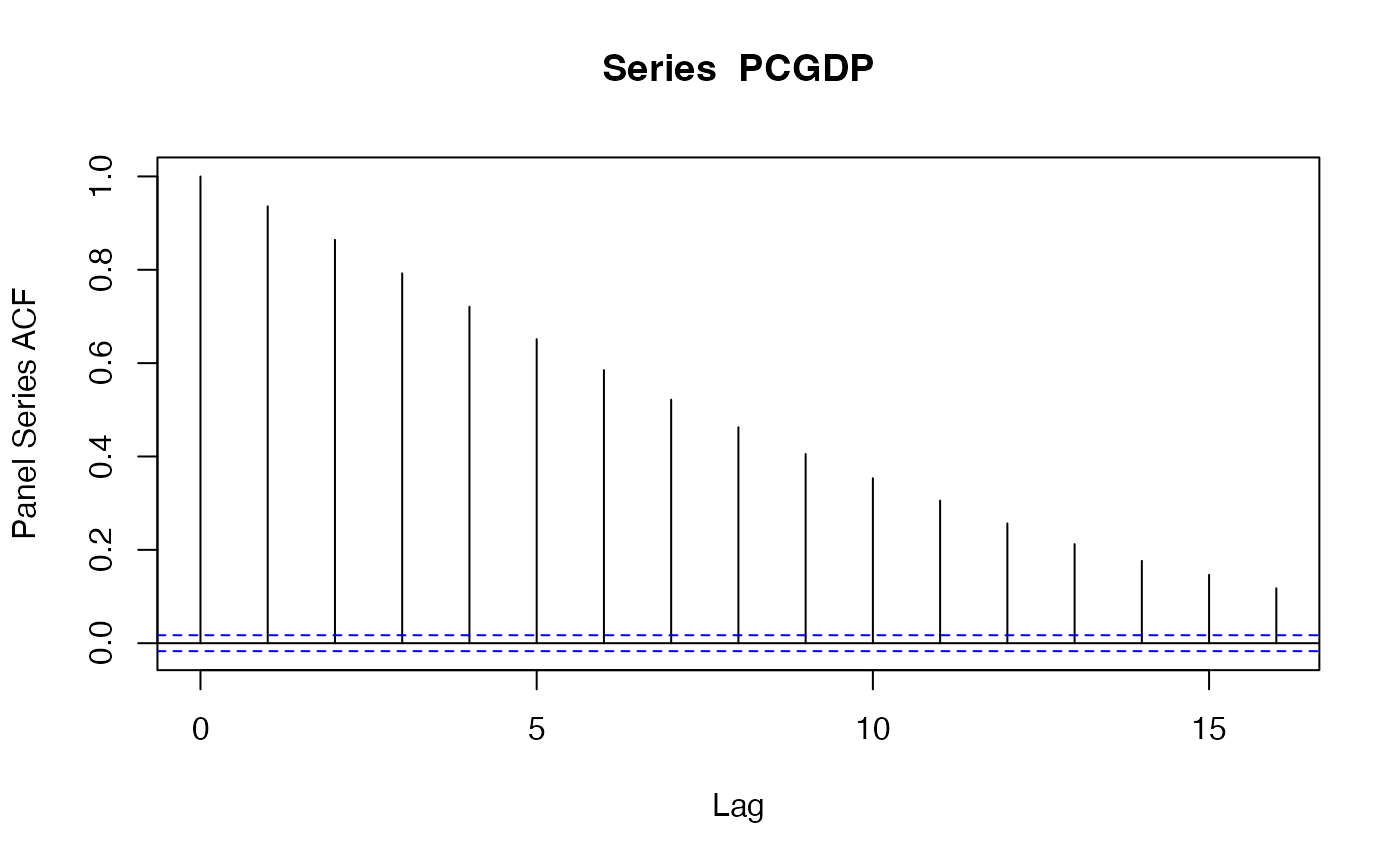
psacf(PCGDP) # Same as above, more parsimonious
## Using indexed data:
wldi <- findex_by(wlddev, iso3c, year) # Creating a indexed frame
PCGDP <- wldi$PCGDP # Indexed Series of GDP per Capita
LIFEEX <- wldi$LIFEEX # Indexed Series of Life Expectancy
psacf(PCGDP) # Same as above, more parsimonious
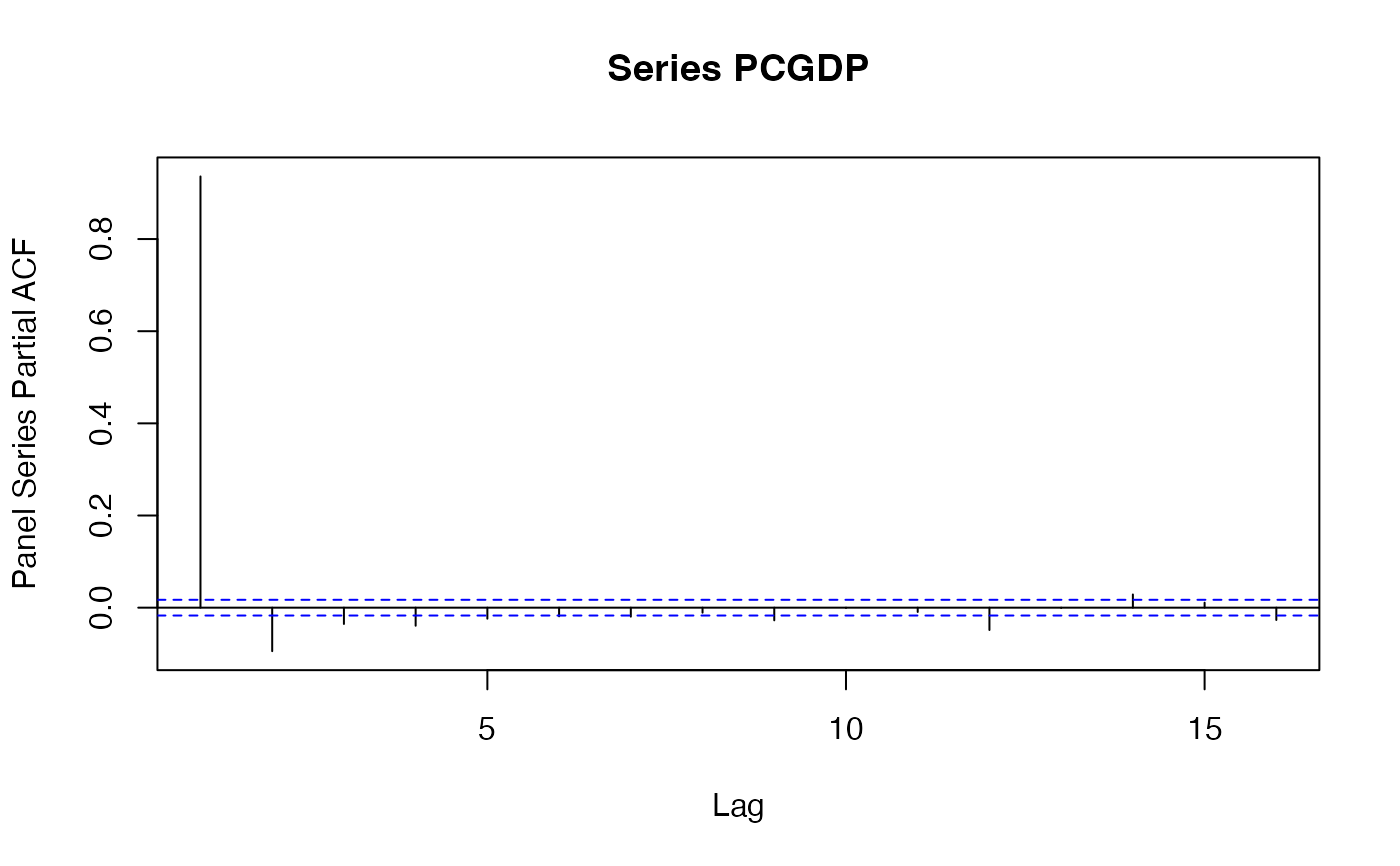
 pspacf(PCGDP)
pspacf(PCGDP)
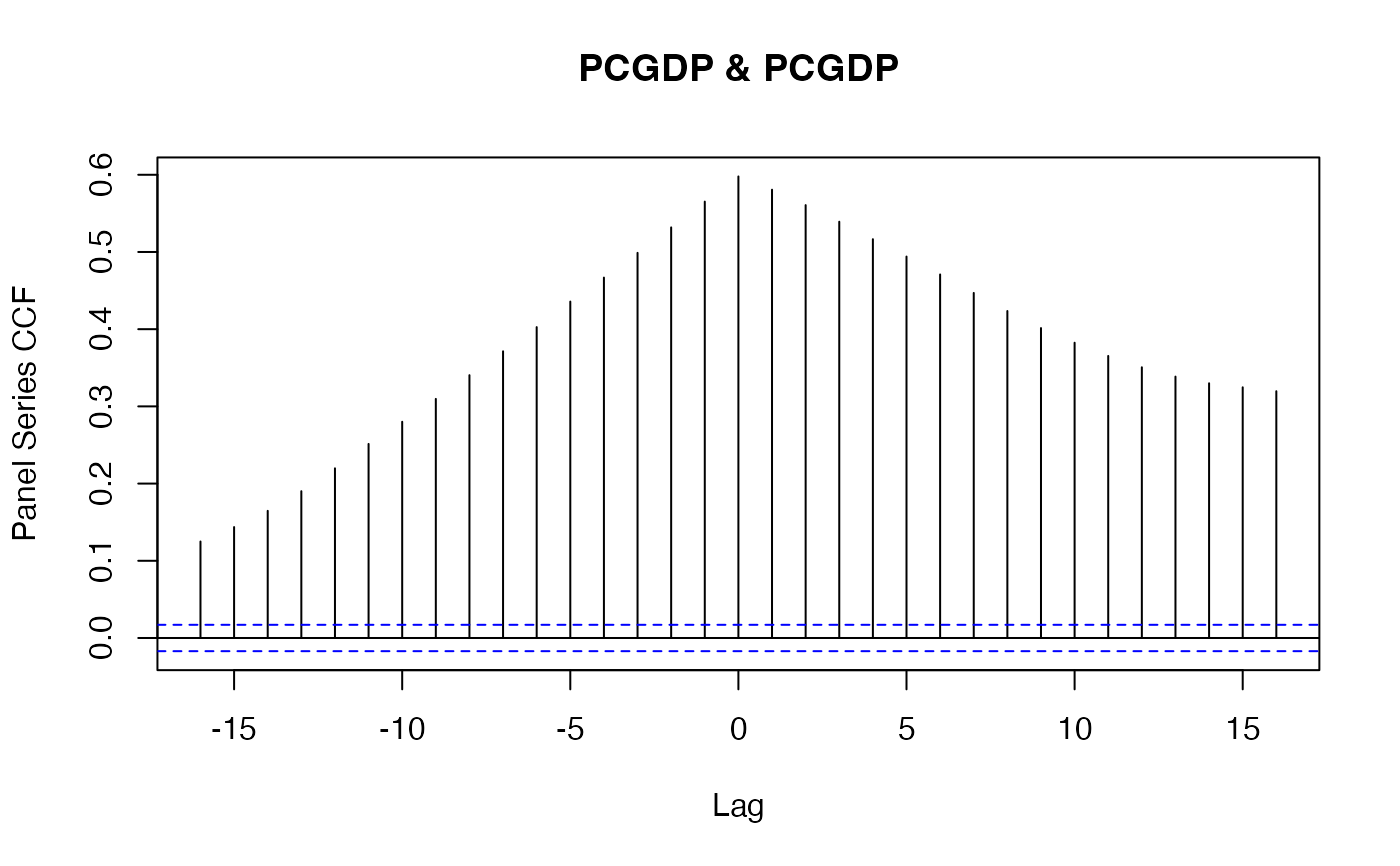
 psccf(PCGDP, LIFEEX)
psccf(PCGDP, LIFEEX)
 psacf(wldi[c(9:10,12)])
psacf(wldi[c(9:10,12)])
 pspacf(wldi[c(9:10,12)])
pspacf(wldi[c(9:10,12)])
