In previous posts I introduced collapse, a powerful (C/C++ based) new framework for data transformation and statistical computing in R - providing advanced grouped, weighted, time series, panel data and recursive computations in R at superior execution speeds, greater flexibility and programmability.
collapse 1.4 released this week additionally introduces an enhanced attribute handling system which enables non-destructive manipulation of vector, matrix or data frame based objects in R. With this post I aim to briefly introduce this attribute handling system and demonstrate that:
collapse non-destructively handles all major matrix (time series) and data frame based classes in R.
Using collapse functions on these objects yields uniform handling at higher computation speeds.
Data Frame Based Objects
The three major data frame based classes in R are the base R data.frame, the data.table and the tibble, for which there also exists grouped (dplyr) and time based (tsibble, tibbletime) versions. Additional notable classes are the panel data frame (plm) and the spatial features data frame (sf).
For the former three collapse offer extremely fast and versatile converters qDF, qDT and qTBL that can be used to turn many R objects into data.frame’s, data.table’s or tibble’s, respectively:
library(collapse); library(data.table); library(tibble)
options(datatable.print.nrows = 10,
datatable.print.topn = 2)
identical(qDF(mtcars), mtcars)
## [1] TRUE
mtcarsDT <- qDT(mtcars, row.names.col = "car")
mtcarsDT
## car mpg cyl disp hp drat wt qsec vs am gear carb
## 1: Mazda RX4 21.0 6 160 110 3.90 2.620 16.46 0 1 4 4
## 2: Mazda RX4 Wag 21.0 6 160 110 3.90 2.875 17.02 0 1 4 4
## ---
## 31: Maserati Bora 15.0 8 301 335 3.54 3.570 14.60 0 1 5 8
## 32: Volvo 142E 21.4 4 121 109 4.11 2.780 18.60 1 1 4 2
mtcarsTBL <- qTBL(mtcars, row.names.col = "car")
print(mtcarsTBL, n = 3)
## # A tibble: 32 x 12
## car mpg cyl disp hp drat wt qsec vs am gear carb
## <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 Mazda RX4 21 6 160 110 3.9 2.62 16.5 0 1 4 4
## 2 Mazda RX4 Wag 21 6 160 110 3.9 2.88 17.0 0 1 4 4
## 3 Datsun 710 22.8 4 108 93 3.85 2.32 18.6 1 1 4 1
## # ... with 29 more rowsThese objects can then be manipulated using an advanced and attribute preserving set of (S3 generic) statistical and data manipulation functions. The following infographic summarizes the core collapse namespace:
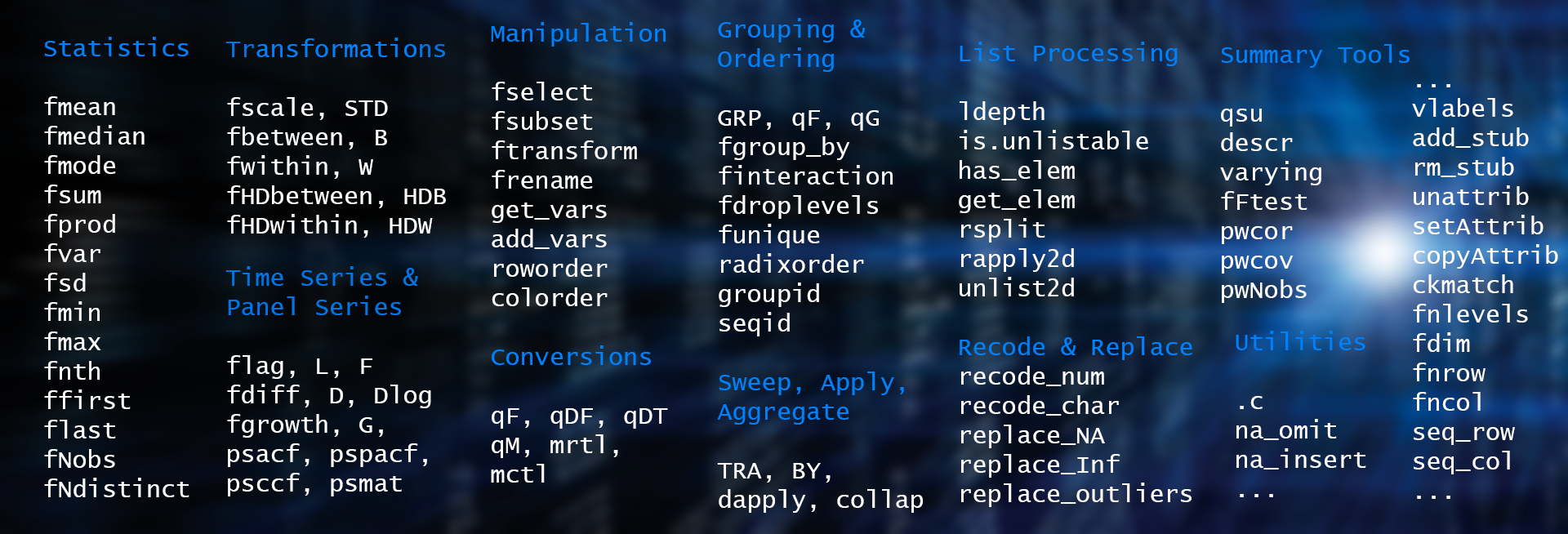
More details are provided in the freshly released cheat sheet, and further in the documentation and vignettes.
The statistical functions internally handle grouped and / or weighted computations on vectors, matrices and data frames, and seek to keep the attributes of the object.
# Simple data frame: Grouped mean by cyl -> groups = row.names
fmean(fselect(mtcars, mpg, disp, drat), g = mtcars$cyl)
## mpg disp drat
## 4 26.66364 105.1364 4.070909
## 6 19.74286 183.3143 3.585714
## 8 15.10000 353.1000 3.229286With fgroup_by, collapse also introduces a fast grouping mechanism that works together with grouped_df versions of all statistical and transformation functions:
# Using Pipe operators and grouped data frames
library(magrittr)
mtcars %>% fgroup_by(cyl) %>%
fselect(mpg, disp, drat, wt) %>% fmean
## cyl mpg disp drat wt
## 1 4 26.66364 105.1364 4.070909 2.285727
## 2 6 19.74286 183.3143 3.585714 3.117143
## 3 8 15.10000 353.1000 3.229286 3.999214
# This is still a data.table
mtcarsDT %>% fgroup_by(cyl) %>%
fselect(mpg, disp, drat, wt) %>% fmean
## cyl mpg disp drat wt
## 1: 4 26.66364 105.1364 4.070909 2.285727
## 2: 6 19.74286 183.3143 3.585714 3.117143
## 3: 8 15.10000 353.1000 3.229286 3.999214
# Same with tibble: here computing weighted group means -> also saves sum of weights in each group
mtcarsTBL %>% fgroup_by(cyl) %>%
fselect(mpg, disp, drat, wt) %>% fmean(wt)
## # A tibble: 3 x 5
## cyl sum.wt mpg disp drat
## <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 4 25.1 25.9 110. 4.03
## 2 6 21.8 19.6 185. 3.57
## 3 8 56.0 14.8 362. 3.21A specialty of the grouping mechanism is that it fully preserves the structure / attributes of the object, and thus permits the creation of a grouped version of any data frame like object.
# This created a grouped data.table
gmtcarsDT <- mtcarsDT %>% fgroup_by(cyl)
gmtcarsDT
## car mpg cyl disp hp drat wt qsec vs am gear carb
## 1: Mazda RX4 21.0 6 160 110 3.90 2.620 16.46 0 1 4 4
## 2: Mazda RX4 Wag 21.0 6 160 110 3.90 2.875 17.02 0 1 4 4
## ---
## 31: Maserati Bora 15.0 8 301 335 3.54 3.570 14.60 0 1 5 8
## 32: Volvo 142E 21.4 4 121 109 4.11 2.780 18.60 1 1 4 2
##
## Grouped by: cyl [3 | 11 (3.5)]
# The print shows: [N. groups | Avg. group size (SD around avg. group size)]
# Subsetting drops groups
gmtcarsDT[1:2]
## car mpg cyl disp hp drat wt qsec vs am gear carb
## 1: Mazda RX4 21 6 160 110 3.9 2.620 16.46 0 1 4 4
## 2: Mazda RX4 Wag 21 6 160 110 3.9 2.875 17.02 0 1 4 4
# Any class-specific methods are independent of the attached groups
gmtcarsDT[, new := mean(mpg)]
gmtcarsDT[, lapply(.SD, mean), by = vs, .SDcols = -1L] # Again groups are dropped
## vs mpg cyl disp hp drat wt qsec am gear carb
## 1: 0 16.61667 7.444444 307.1500 189.72222 3.392222 3.688556 16.69389 0.3333333 3.555556 3.611111
## 2: 1 24.55714 4.571429 132.4571 91.35714 3.859286 2.611286 19.33357 0.5000000 3.857143 1.785714
## new
## 1: 20.09062
## 2: 20.09062
# Groups are always preserved in column-subsetting operations
gmtcarsDT[, 9:13]
## vs am gear carb new
## 1: 0 1 4 4 20.09062
## 2: 0 1 4 4 20.09062
## ---
## 31: 0 1 5 8 20.09062
## 32: 1 1 4 2 20.09062
##
## Grouped by: cyl [3 | 11 (3.5)]The grouping is also dropped in aggregations, but preserved in transformations keeping data dimensions:
# Grouped medians
fmedian(gmtcarsDT[, 9:13])
## cyl vs am gear carb new
## 1: 4 1 1 4 2.0 20.09062
## 2: 6 1 0 4 4.0 20.09062
## 3: 8 0 0 3 3.5 20.09062
# Note: unique grouping columns are stored in the attached grouping object
# and added if keep.group_vars = TRUE (the default)
# Replacing data by grouped median (grouping columns are not selected and thus not present)
fmedian(gmtcarsDT[, 4:5], TRA = "replace")
## disp hp
## 1: 167.6 110.0
## 2: 167.6 110.0
## ---
## 31: 350.5 192.5
## 32: 108.0 91.0
##
## Grouped by: cyl [3 | 11 (3.5)]
# Weighted scaling and centering data (here also selecting grouping column)
mtcarsDT %>% fgroup_by(cyl) %>%
fselect(cyl, mpg, disp, drat, wt) %>% fscale(wt)
## cyl wt mpg disp drat
## 1: 6 2.620 0.96916875 -0.6376553 0.7123846
## 2: 6 2.875 0.96916875 -0.6376553 0.7123846
## ---
## 31: 8 3.570 0.07335466 -0.8685527 0.9844833
## 32: 4 2.780 -1.06076989 0.3997723 0.2400387
##
## Grouped by: cyl [3 | 11 (3.5)]As mentioned, this works for any data frame like object, even a suitable list:
# Here computing a weighted grouped standard deviation
as.list(mtcars) %>% fgroup_by(cyl, vs, am) %>%
fsd(wt) %>% str
## List of 11
## $ cyl : num [1:7] 4 4 4 6 6 8 8
## $ vs : num [1:7] 0 1 1 0 1 0 0
## $ am : num [1:7] 1 0 1 1 0 0 1
## $ sum.wt: num [1:7] 2.14 8.8 14.2 8.27 13.55 ...
## $ mpg : num [1:7] 0 1.236 4.833 0.655 1.448 ...
## $ disp : num [1:7] 0 11.6 19.25 7.55 39.93 ...
## $ hp : num [1:7] 0 17.3 22.7 32.7 8.3 ...
## $ drat : num [1:7] 0 0.115 0.33 0.141 0.535 ...
## $ qsec : num [1:7] 0 1.474 0.825 0.676 0.74 ...
## $ gear : num [1:7] 0 0.477 0.32 0.503 0.519 ...
## $ carb : num [1:7] 0 0.477 0.511 1.007 1.558 ...
## - attr(*, "row.names")= int [1:7] 1 2 3 4 5 6 7The function fungroup can be used to undo any grouping operation.
identical(mtcarsDT,
mtcarsDT %>% fgroup_by(cyl, vs, am) %>% fungroup)
## [1] TRUEApart from the grouping mechanism with fgroup_by, which is very fast and versatile, collapse also supports regular grouped tibbles created with dplyr:
library(dplyr)
# Same as summarize_all(sum) and considerably faster
mtcars %>% group_by(cyl) %>% fsum
## # A tibble: 3 x 11
## cyl mpg disp hp drat wt qsec vs am gear carb
## <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 4 293. 1157. 909 44.8 25.1 211. 10 8 45 17
## 2 6 138. 1283. 856 25.1 21.8 126. 4 3 27 24
## 3 8 211. 4943. 2929 45.2 56.0 235. 0 2 46 49
# Same as muatate_all(sum)
mtcars %>% group_by(cyl) %>% fsum(TRA = "replace_fill") %>% head(3)
## # A tibble: 3 x 11
## # Groups: cyl [2]
## cyl mpg disp hp drat wt qsec vs am gear carb
## <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 6 138. 1283. 856 25.1 21.8 126. 4 3 27 24
## 2 6 138. 1283. 856 25.1 21.8 126. 4 3 27 24
## 3 4 293. 1157. 909 44.8 25.1 211. 10 8 45 17One major goal of the package is to make R suitable for (large) panel data, thus collapse also supports panel-data.frames created with the plm package:
library(plm)
pwlddev <- pdata.frame(wlddev, index = c("iso3c", "year"))
# Centering (within-transforming) columns 9-12 using the within operator W()
head(W(pwlddev, cols = 9:12), 3)
## iso3c year W.PCGDP W.LIFEEX W.GINI W.ODA
## ABW-1960 ABW 1960 NA -6.547351 NA NA
## ABW-1961 ABW 1961 NA -6.135351 NA NA
## ABW-1962 ABW 1962 NA -5.765351 NA NA
# Computing growth rates of columns 9-12 using the growth operator G()
head(G(pwlddev, cols = 9:12), 3)
## iso3c year G1.PCGDP G1.LIFEEX G1.GINI G1.ODA
## ABW-1960 ABW 1960 NA NA NA NA
## ABW-1961 ABW 1961 NA 0.6274558 NA NA
## ABW-1962 ABW 1962 NA 0.5599782 NA NAPerhaps a note about operators is necessary here before proceeding: collapse offers a set of transformation operators for its vector-valued fast functions:
# Operators
.OPERATOR_FUN
## [1] "STD" "B" "W" "HDB" "HDW" "L" "F" "D" "Dlog" "G"
# Corresponding (programmers) functions
setdiff(.FAST_FUN, .FAST_STAT_FUN)
## [1] "fscale" "fbetween" "fwithin" "fHDbetween" "fHDwithin" "flag" "fdiff"
## [8] "fgrowth"These operators are principally just function shortcuts that exist for parsimony and in-formula use (e.g. to specify dynamic or fixed effects models using lm(), see the documentation). They however also have some useful extra features in the data.frame method, such as internal column-subsetting using the cols argument or stub-renaming transformed columns (adding a ‘W.’ or ‘Gn.’ prefix as shown above). They also permit grouping variables to be passed using formulas, including options to keep (default) or drop those variables in the output. We will see this feature when using time series below.
To round things off for data frames, I demonstrate the use of collapse with classes it was not directly built to support but can also handle very well. Through it’s built in capabilities for handling panel data, tsibble’s can seamlessly be utilized:
library(tsibble)
tsib <- as_tsibble(EuStockMarkets)
# Computing daily and annual growth rates on tsibble
head(G(tsib, c(1, 260), by = ~ key, t = ~ index), 3)
## # A tsibble: 3 x 4 [1s] <UTC>
## # Key: key [1]
## key index G1.value L260G1.value
## <chr> <dttm> <dbl> <dbl>
## 1 DAX 1991-07-01 02:18:33 NA NA
## 2 DAX 1991-07-02 12:00:00 -0.928 NA
## 3 DAX 1991-07-03 21:41:27 -0.441 NA
# Computing a compounded annual growth rate
head(G(tsib, 260, by = ~ key, t = ~ index, power = 1/260), 3)
## # A tsibble: 3 x 3 [1s] <UTC>
## # Key: key [1]
## key index L260G1.value
## <chr> <dttm> <dbl>
## 1 DAX 1991-07-01 02:18:33 NA
## 2 DAX 1991-07-02 12:00:00 NA
## 3 DAX 1991-07-03 21:41:27 NASimilarly for tibbletime:
library(tibbletime); library(tsbox)
# Using the tsbox converter
tibtm <- ts_tibbletime(EuStockMarkets)
# Computing daily and annual growth rates on tibbletime
head(G(tibtm, c(1, 260), t = ~ time), 3)
## # A time tibble: 3 x 9
## # Index: time
## time G1.DAX L260G1.DAX G1.SMI L260G1.SMI G1.CAC L260G1.CAC G1.FTSE L260G1.FTSE
## <dttm> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 1991-07-01 02:18:27 NA NA NA NA NA NA NA NA
## 2 1991-07-02 12:01:32 -0.928 NA 0.620 NA -1.26 NA 0.679 NA
## 3 1991-07-03 21:44:38 -0.441 NA -0.586 NA -1.86 NA -0.488 NA
# ...Finally lets consider the simple features data frame:
library(sf)
nc <- st_read(system.file("shape/nc.shp", package="sf"))
## Reading layer `nc' from data source `C:\Users\Sebastian Krantz\Documents\R\win-library\4.0\sf\shape\nc.shp' using driver `ESRI Shapefile'
## Simple feature collection with 100 features and 14 fields
## geometry type: MULTIPOLYGON
## dimension: XY
## bbox: xmin: -84.32385 ymin: 33.88199 xmax: -75.45698 ymax: 36.58965
## geographic CRS: NAD27
# Fast selecting columns (need to add 'geometry' column to not break the class)
plot(fselect(nc, AREA, geometry))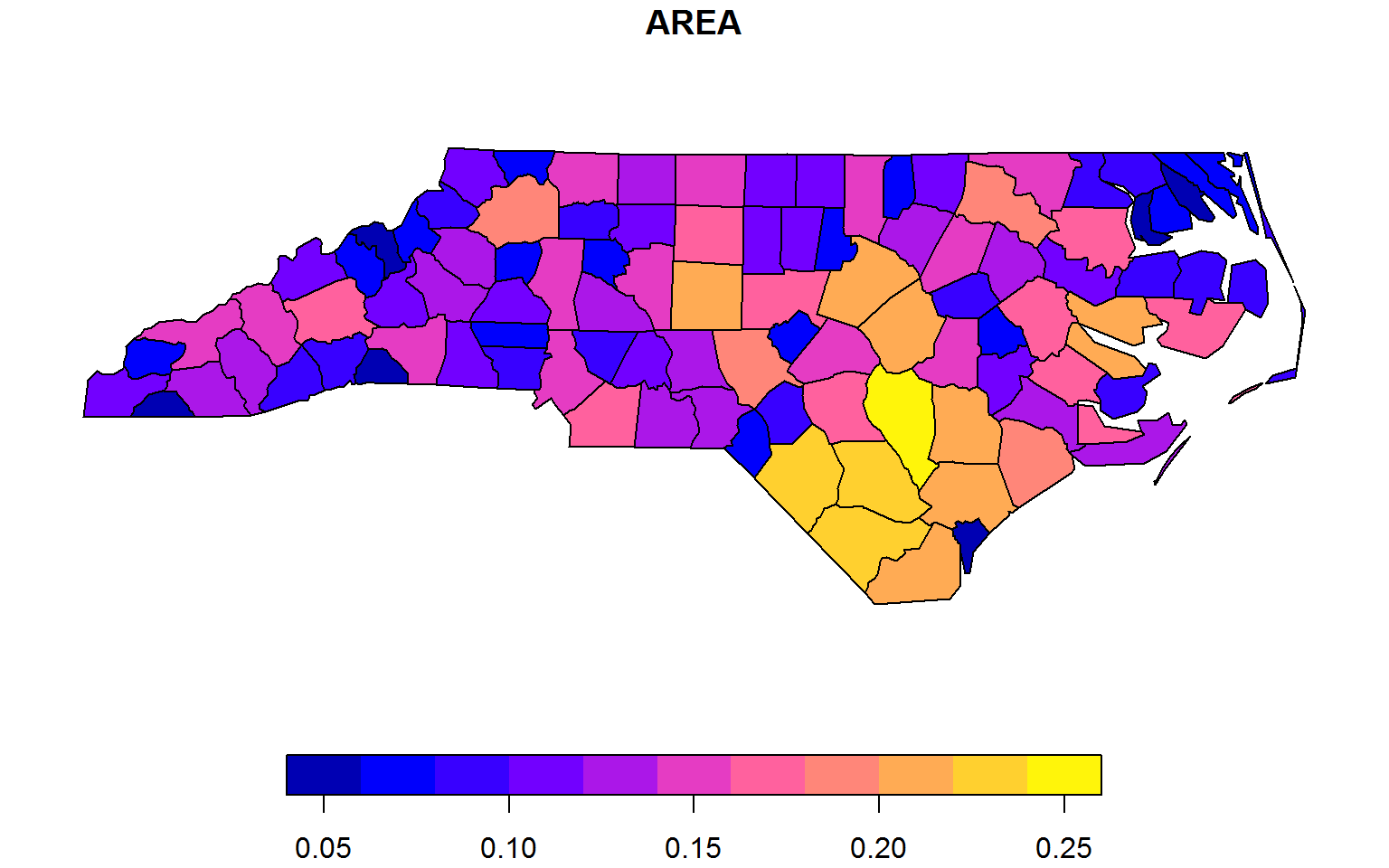
# Subsetting
fsubset(nc, AREA > 0.23, NAME, AREA, geometry)
## Simple feature collection with 3 features and 2 fields
## geometry type: MULTIPOLYGON
## dimension: XY
## bbox: xmin: -84.32385 ymin: 33.88199 xmax: -75.45698 ymax: 36.58965
## geographic CRS: NAD27
## NAME AREA geometry
## 1 Sampson 0.241 MULTIPOLYGON (((-78.11377 3...
## 2 Robeson 0.240 MULTIPOLYGON (((-78.86451 3...
## 3 Columbus 0.240 MULTIPOLYGON (((-78.65572 3...
# Standardizing numeric columns (by reference)
settransformv(nc, is.numeric, STD, apply = FALSE)
# Note: Here using using operator STD() instead of fscale() to stub-rename standardized columns.
# apply = FALSE uses STD.data.frame on all numeric columns instead of lapply(data, STD)
head(nc, 2)
## Simple feature collection with 2 features and 26 fields
## geometry type: MULTIPOLYGON
## dimension: XY
## bbox: xmin: -81.74107 ymin: 36.23436 xmax: -80.90344 ymax: 36.58965
## geographic CRS: NAD27
## AREA PERIMETER CNTY_ CNTY_ID NAME FIPS FIPSNO CRESS_ID BIR74 SID74 NWBIR74 BIR79 SID79
## 1 0.114 1.442 1825 1825 Ashe 37009 37009 5 1091 1 10 1364 0
## 2 0.061 1.231 1827 1827 Alleghany 37005 37005 3 487 0 10 542 3
## NWBIR79 geometry STD.AREA STD.PERIMETER STD.CNTY_ STD.CNTY_ID STD.FIPSNO
## 1 19 MULTIPOLYGON (((-81.47276 3... -0.249186 -0.4788595 -1.511125 -1.511125 -1.568344
## 2 12 MULTIPOLYGON (((-81.23989 3... -1.326418 -0.9163351 -1.492349 -1.492349 -1.637282
## STD.CRESS_ID STD.BIR74 STD.SID74 STD.NWBIR74 STD.BIR79 STD.SID79 STD.NWBIR79
## 1 -1.568344 -0.5739411 -0.7286824 -0.7263602 -0.5521659 -0.8863574 -0.6750055
## 2 -1.637282 -0.7308990 -0.8571979 -0.7263602 -0.7108697 -0.5682866 -0.6785480Matrix Based Objects
collapse also offers a converter qM to efficiently convert various objects to matrix:
m <- qM(mtcars)Grouped and / or weighted computations and transformations work as with with data frames:
# Grouped means
fmean(m, g = mtcars$cyl)
## mpg cyl disp hp drat wt qsec vs am gear carb
## 4 26.66364 4 105.1364 82.63636 4.070909 2.285727 19.13727 0.9090909 0.7272727 4.090909 1.545455
## 6 19.74286 6 183.3143 122.28571 3.585714 3.117143 17.97714 0.5714286 0.4285714 3.857143 3.428571
## 8 15.10000 8 353.1000 209.21429 3.229286 3.999214 16.77214 0.0000000 0.1428571 3.285714 3.500000
# Grouped and weighted standardizing
head(fscale(m, g = mtcars$cyl, w = mtcars$wt), 3)
## mpg cyl disp hp drat wt qsec vs
## Mazda RX4 0.9691687 NaN -0.63765527 -0.5263758 0.7123846 -1.6085211 -1.0438559 -1.2509539
## Mazda RX4 Wag 0.9691687 NaN -0.63765527 -0.5263758 0.7123846 -0.8376064 -0.6921302 -1.2509539
## Datsun 710 -0.7333024 NaN -0.08822497 0.4896429 -0.5526066 -0.1688057 -0.4488514 0.2988833
## am gear carb
## Mazda RX4 1.250954 0.27612029 0.386125
## Mazda RX4 Wag 1.250954 0.27612029 0.386125
## Datsun 710 0.719370 -0.09429567 -1.133397Various matrix-based time series classes such as xts / zoo and timeSeries are also easily handled:
# ts / mts
# Note: G() by default renames the columns, fgrowth() does not
plot(G(EuStockMarkets))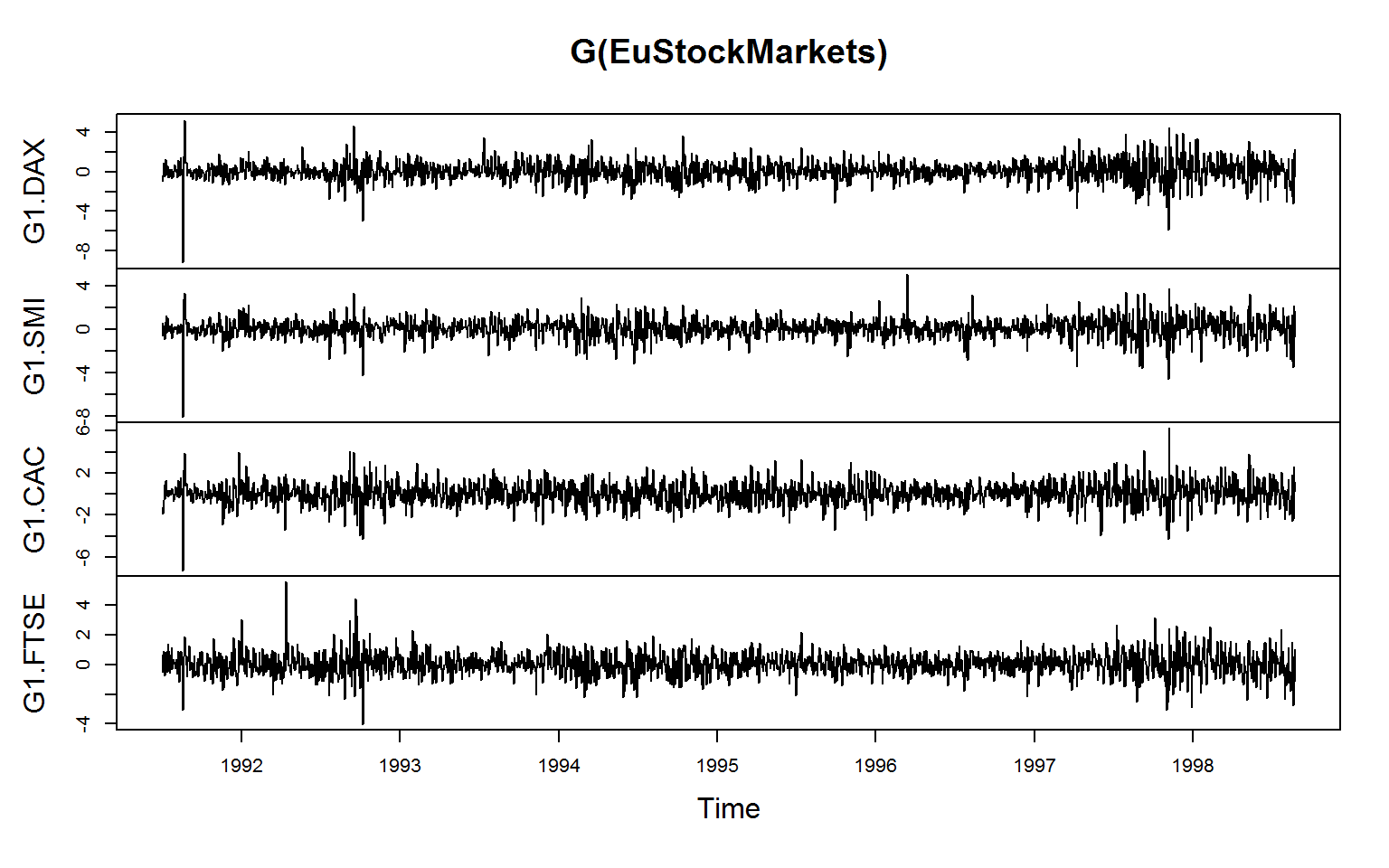
# xts
library(xts)
ESM_xts <- ts_xts(EuStockMarkets) # using tsbox::ts_xts
head(G(ESM_xts), 3)
## G1.DAX G1.SMI G1.CAC G1.FTSE
## 1991-07-01 02:18:27 NA NA NA NA
## 1991-07-02 12:01:32 -0.9283193 0.6197485 -1.257897 0.6793256
## 1991-07-03 21:44:38 -0.4412412 -0.5863192 -1.856612 -0.4877652
plot(G(ESM_xts), legend.loc = "bottomleft")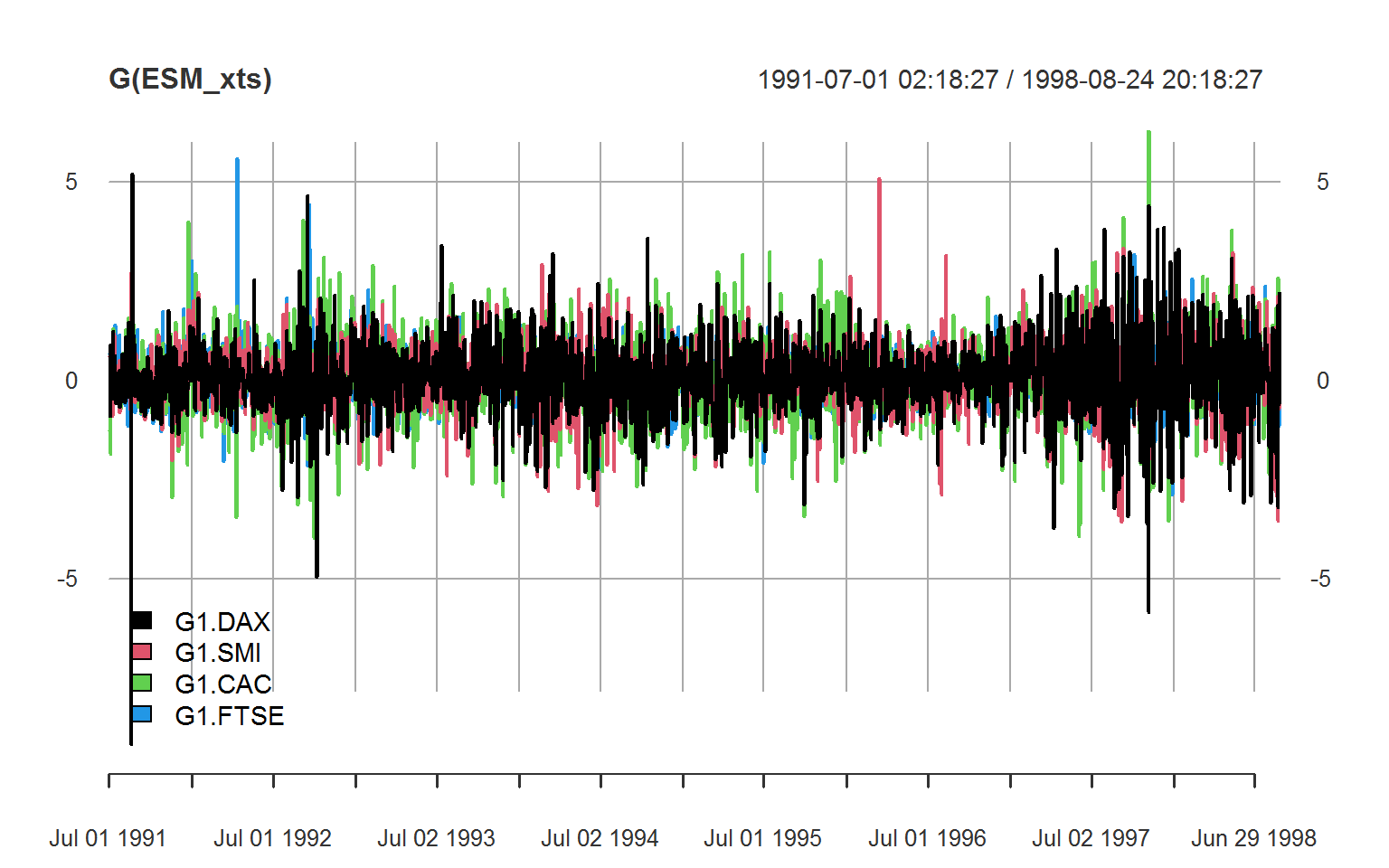
# timeSeries
library(timeSeries) # using tsbox::ts_timeSeries
ESM_timeSeries <- ts_timeSeries(EuStockMarkets)
# Note: G() here also renames the columns but the names of the series are also stored in an attribute
head(G(ESM_timeSeries), 3)
## GMT
## DAX SMI CAC FTSE
## 1991-06-30 23:18:27 NA NA NA NA
## 1991-07-02 09:01:32 -0.9283193 0.6197485 -1.257897 0.6793256
## 1991-07-03 18:44:38 -0.4412412 -0.5863192 -1.856612 -0.4877652
plot(G(ESM_timeSeries), plot.type = "single", at = "pretty")
legend("bottomleft", colnames(G(qM(ESM_timeSeries))), lty = 1, col = 1:4)
Aggregating these objects yields a plain matrix with groups in the row-names:
# Aggregating by year: creates plain matrix with row-names (g is second argument)
EuStockMarkets %>% fmedian(round(time(.)))
## DAX SMI CAC FTSE
## 1991 1628.750 1678.10 1772.8 2443.60
## 1992 1649.550 1733.30 1863.5 2558.50
## 1993 1606.640 2061.70 1837.5 2773.40
## 1994 2089.770 2727.10 2148.0 3111.40
## 1995 2072.680 2591.60 1918.5 3091.70
## 1996 2291.820 3251.60 1946.2 3661.65
## 1997 2861.240 3906.55 2297.1 4075.35
## 1998 4278.725 6077.40 3002.7 5222.20
## 1999 5905.150 8102.70 4205.4 5884.50
# Same thing with the other objects
all_obj_equal(ESM_xts %>% fmedian(substr(time(.), 1L, 4L)),
ESM_timeSeries %>% fmedian(substr(time(.), 1L, 4L)))
## [1] TRUEBenchmarks
Extensive benchmarks and examples against native dplyr / tibble and plm are provided here and here, making it evident that collapse provides both greater versatility and massive performance improvements over the methods defined for these objects. Benchmarks against data.table were provided in a previous post, where collapse compared favorably on a 2-core machine (particularly for weighted and := type operations). In general collapse functions are extremely well optimized, with basic execution speeds below 30 microseconds, and efficiently scale to larger operations. Most importantly, they preserve the data structure and attributes (including column attributes) of the objects passed to them. They also efficiently skip missing values and avoid some of the undesirable behavior endemic of base R1.
Here I will add to the above resources just a small benchmark to prove that computations with collapse are also faster than any native methods and suggested programming principles for the various time series classes:
library(dplyr) # needed for tibbletime / tsibble comparison
library(microbenchmark)
# Computing the first difference
microbenchmark(ts = diff(EuStockMarkets),
collapse_ts = fdiff(EuStockMarkets),
xts = diff(ESM_xts),
collapse_xts = fdiff(ESM_xts),
timeSeries = diff(ESM_timeSeries),
collapse_timeSeries = fdiff(ESM_timeSeries),
# taking difference function from tsibble
dplyr_tibbletime = mutate_at(tibtm, 2:5, difference, order_by = tibtm$time),
collapse_tibbletime_D = D(tibtm, t = ~ time),
# collapse equivalent to the dplyr method (tfmv() abbreviates ftransformv())
collapse_tibbletime_tfmv = tfmv(tibtm, 2:5, fdiff, t = time, apply = FALSE),
# dplyr helpers provided by tsibble package
dplyr_tsibble = mutate(group_by_key(tsib), value = difference(value, order_by = index)),
collapse_tsibble_D = D(tsib, 1, 1, ~ key, ~ index),
# Again we can do the same using collapse (tfm() abbreviates ftransform())
collapse_tsibble_tfm = tfm(tsib, value = fdiff(value, 1, 1, key, index)))
## Unit: microseconds
## expr min lq mean median uq max neval cld
## ts 1681.018 2061.2220 2719.8758 2313.1290 3055.6870 9473.855 100 a
## collapse_ts 25.883 56.4505 163.3896 70.9540 88.1340 8732.636 100 a
## xts 93.713 176.7150 279.5615 200.3655 257.0395 3656.561 100 a
## collapse_xts 42.840 78.0940 116.1231 99.7370 126.5120 597.973 100 a
## timeSeries 1964.386 2287.4700 3185.6565 2742.4210 3310.7180 17093.102 100 a
## collapse_timeSeries 52.658 93.9355 2483.3061 108.4390 132.7590 235259.074 100 a
## dplyr_tibbletime 8097.625 10173.1265 14784.9888 11176.0700 12863.3355 239144.113 100 b
## collapse_tibbletime_D 568.967 630.5490 857.0829 751.2595 850.3270 8017.301 100 a
## collapse_tibbletime_tfmv 501.137 585.4780 773.2819 664.2410 817.0815 2922.035 100 a
## dplyr_tsibble 10386.434 12641.1040 14690.4243 13558.5910 15970.5640 32364.171 100 b
## collapse_tsibble_D 963.897 1056.4935 1338.5404 1210.8955 1477.7525 3665.485 100 a
## collapse_tsibble_tfm 940.246 993.5720 1272.6027 1114.9525 1420.6320 6188.128 100 a
# Sequence of lagged/leaded and iterated differences (not supported by either of these methods)
head(fdiff(ESM_xts, -1:1, diff = 1:2)[, 1:6], 3)
## FD1.DAX FD2.DAX DAX D1.DAX D2.DAX FD1.SMI
## 1991-07-01 02:18:27 15.12 8.00 1628.75 NA NA -10.4
## 1991-07-02 12:01:32 7.12 21.65 1613.63 -15.12 NA 9.9
## 1991-07-03 21:44:38 -14.53 -17.41 1606.51 -7.12 8 -5.5
head(D(tibtm, -1:1, diff = 1:2, t = ~ time), 3)
## # A time tibble: 3 x 21
## # Index: time
## time FD1.DAX FD2.DAX DAX D1.DAX D2.DAX FD1.SMI FD2.SMI SMI D1.SMI D2.SMI FD1.CAC
## <dttm> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 1991-07-01 02:18:27 15.1 8.00 1629. NA NA -10.4 -20.3 1678. NA NA 22.3
## 2 1991-07-02 12:01:32 7.12 21.7 1614. -15.1 NA 9.9 15.4 1688. 10.4 NA 32.5
## 3 1991-07-03 21:44:38 -14.5 -17.4 1607. -7.12 8.00 -5.5 -3 1679. -9.9 -20.3 9.9
## # ... with 9 more variables: FD2.CAC <dbl>, CAC <dbl>, D1.CAC <dbl>, D2.CAC <dbl>, FD1.FTSE <dbl>,
## # FD2.FTSE <dbl>, FTSE <dbl>, D1.FTSE <dbl>, D2.FTSE <dbl>
head(D(tsib, -1:1, diff = 1:2, ~ key, ~ index), 3)
## # A tsibble: 3 x 7 [1s] <UTC>
## # Key: key [1]
## key index FD1.value FD2.value value D1.value D2.value
## <chr> <dttm> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 DAX 1991-07-01 02:18:33 15.1 8.00 1629. NA NA
## 2 DAX 1991-07-02 12:00:00 7.12 21.7 1614. -15.1 NA
## 3 DAX 1991-07-03 21:41:27 -14.5 -17.4 1607. -7.12 8.00
microbenchmark(collapse_xts = fdiff(ESM_xts, -1:1, diff = 1:2),
collapse_tibbletime = D(tibtm, -1:1, diff = 1:2, t = ~ time),
collapse_tsibble = D(tsib, -1:1, diff = 1:2, ~ key, ~ index))
## Unit: microseconds
## expr min lq mean median uq max neval cld
## collapse_xts 147.263 218.2155 358.0477 254.362 290.508 8398.843 100 a
## collapse_tibbletime 750.144 988.4405 1348.9559 1026.149 1085.722 13947.942 100 b
## collapse_tsibble 1258.421 1653.3505 1793.5130 1707.793 1757.327 8411.784 100 cConclusion
This concludes this short demonstration. collapse is an advanced, fast and versatile data manipulation package. If you have followed until here I am convinced you will find it very useful, particularly if you are working in advanced statistics, econometrics, surveys, time series, panel data and the like, or if you care much about performance and non-destructive working in R. For more information about the package see the website, study the cheat sheet or call help("collapse-documentation") after install to bring up the built-in documentation.
Appendix: So how does this all actually work?
Statistical functions like fmean are S3 generic with user visible ‘default’, ‘matrix’ and ‘data.frame’ methods, and hidden ‘list’ and ‘grouped_df’ methods. Transformation functions like fwithin additionally have ‘pseries’ and ‘pdata.frame’ methods to support plm objects.
The ‘default’, ‘matrix’ and ‘data.frame’ methods handle object attributes intelligently. In the case of ‘data.frame’s’ only the ‘row.names’ attribute is modified accordingly, other attributes (including column attributes) are preserved. This also holds for data manipulation functions like fselect, fsubset, ftransform etc.. ‘default’ and ‘matrix’ methods preserve attributes as long as the data dimensions are kept.
In addition, the ‘default’ method checks if its argument is actually a matrix, and calls the matrix method if is.matrix(x) && !inherits(x, "matrix") is TRUE. This prevents classed matrix-based objects (such as xts time series) not inheriting from ‘matrix’ being handled by the default method.
For example.
mean(NA, na.rm = TRUE)givesNaN,sum(NA, na.rm = TRUE)gives0andmax(NA, na.rm = TRUE)gives-Infwhereasall_identical(NA_real_, fmean(NA), fsum(NA), fmax(NA)).na.rm = TRUEis the default setting for all collapse functions. Settingna.rm = FALSEalso checks for missing values and returnsNAif found instead of just running through the entire computation and then returning aNAorNaNvalue which is unreliable and inefficient.↩︎